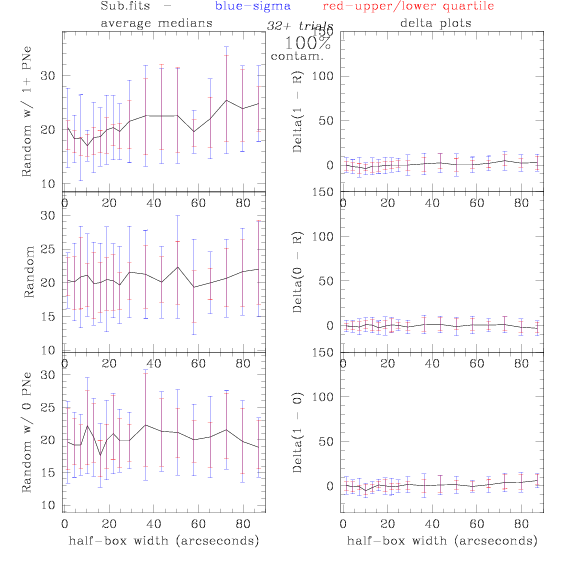
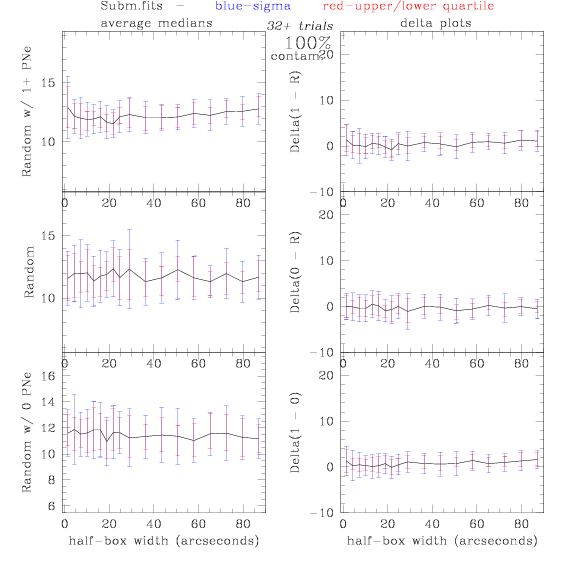
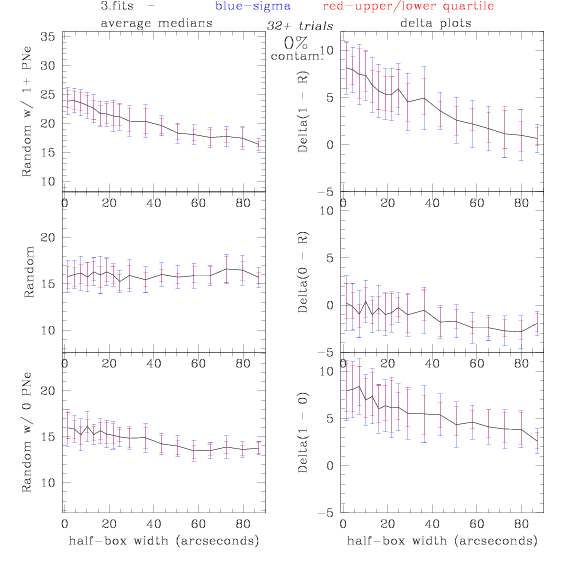
PNe14 - Variable levels of contamination on simulated
data - random boxes
Contamination was
artifically added to our theoretical
catalogs: After
a theoretical catalog was generated, we randomly selected 0% - 100% of
the PNe to replace with random coordinates.
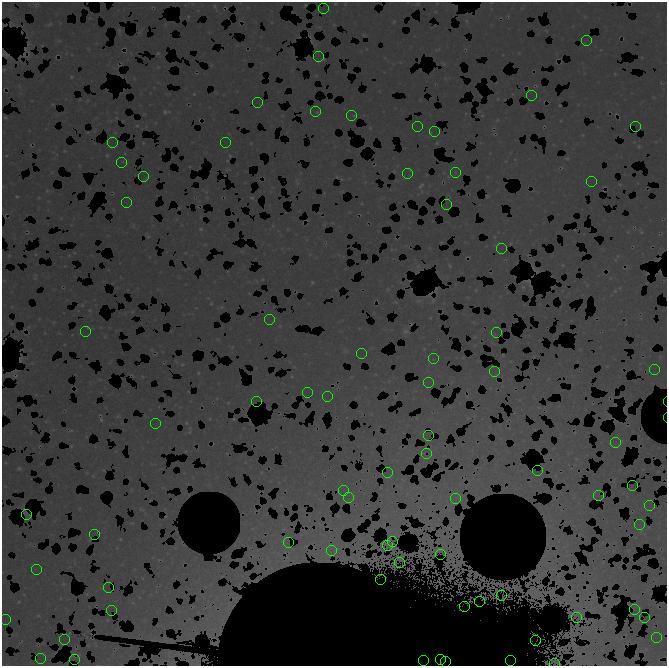
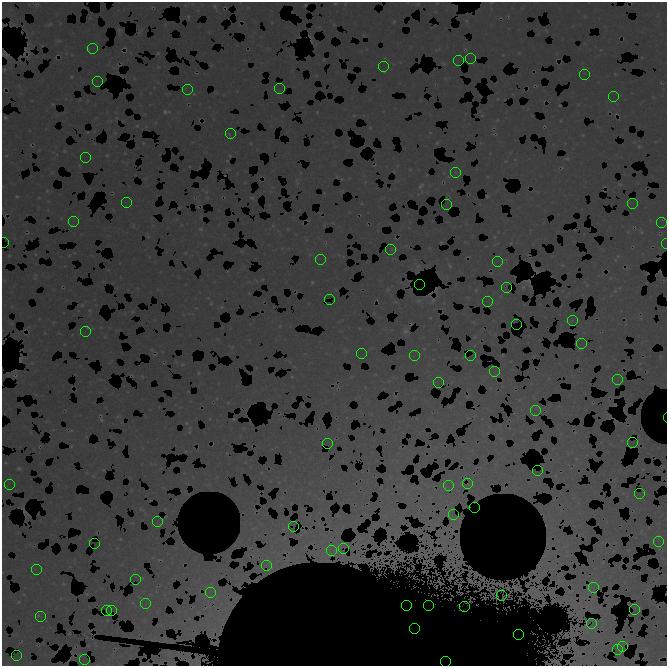
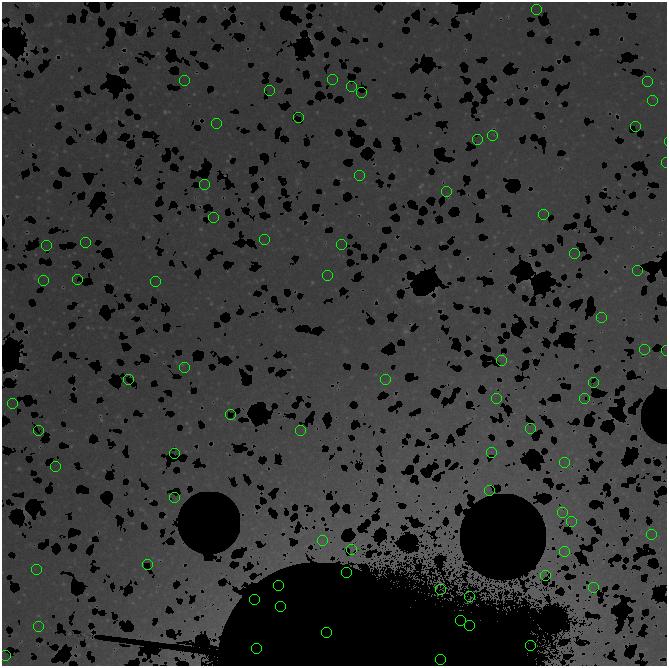
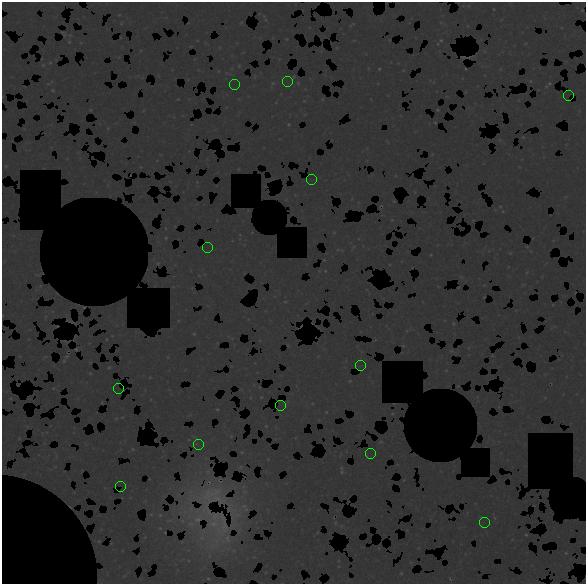
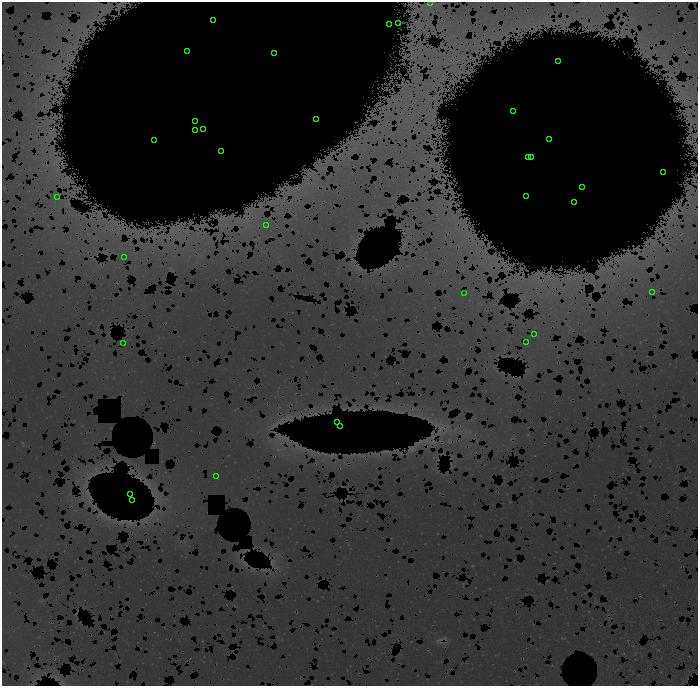
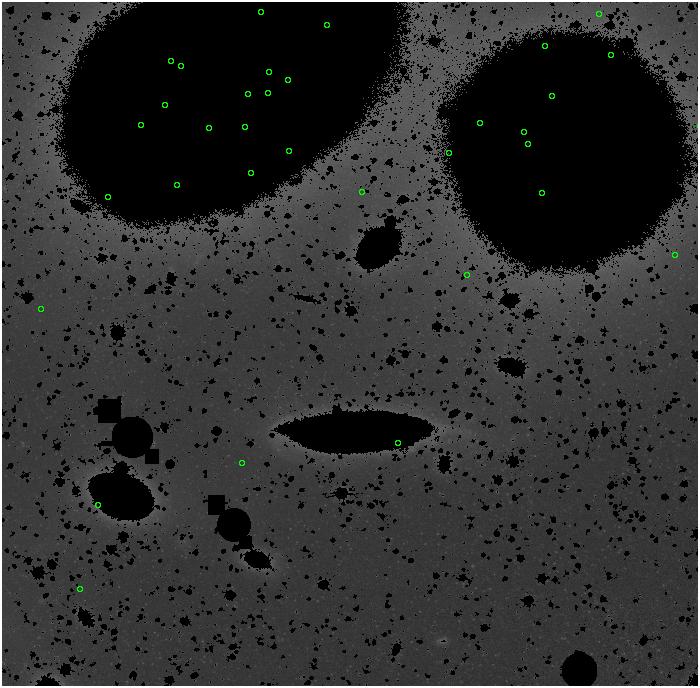
Random box-selection was used to choose
regions for comparison.
Possible method problems:
---only one randomised catalog is used for each Monte-Carlo set of
trials. Perhaps the subsampling should be done for each trial... and it
is! in this version.
Contamination level links:
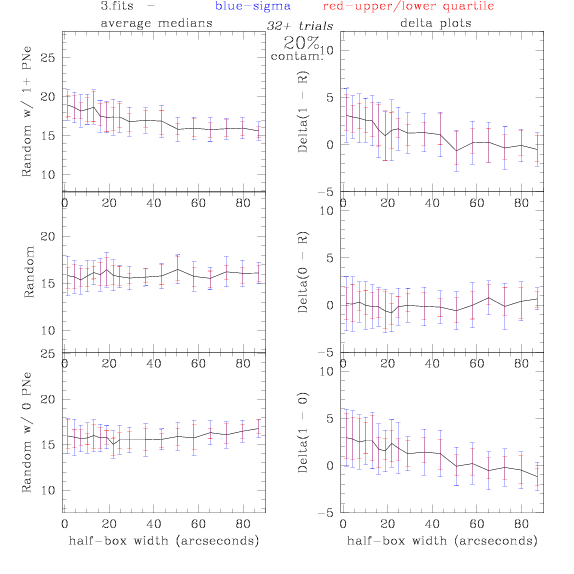
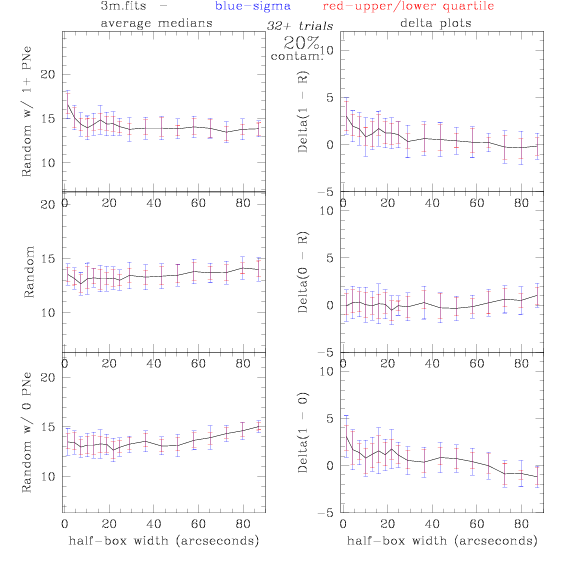
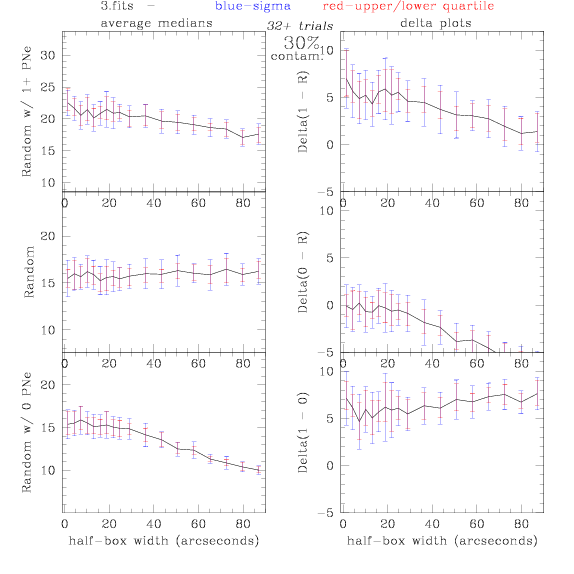
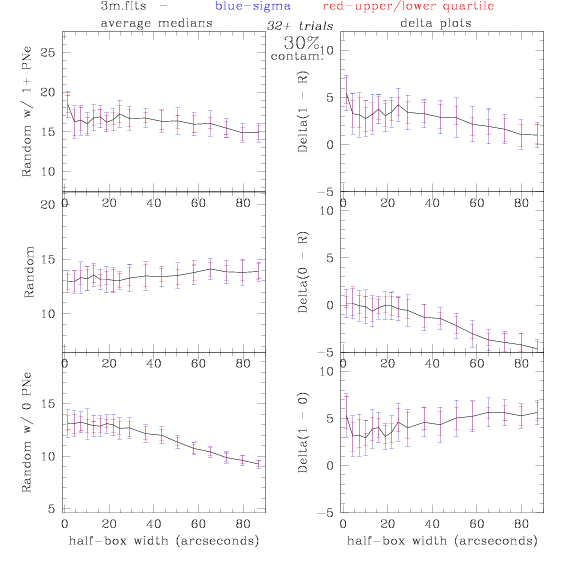
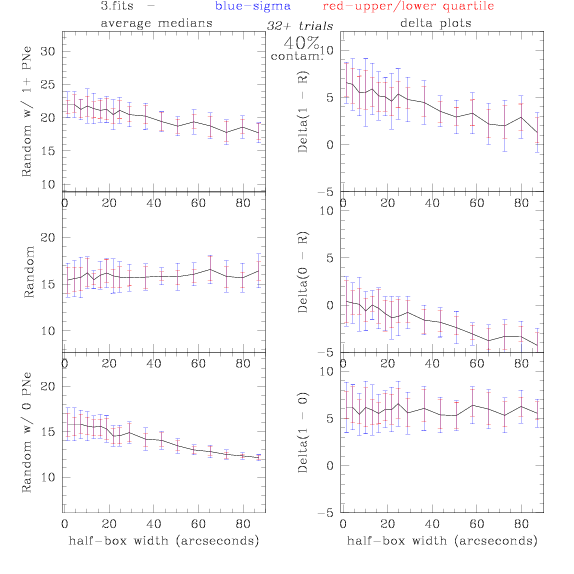
3.fits/3m.fits

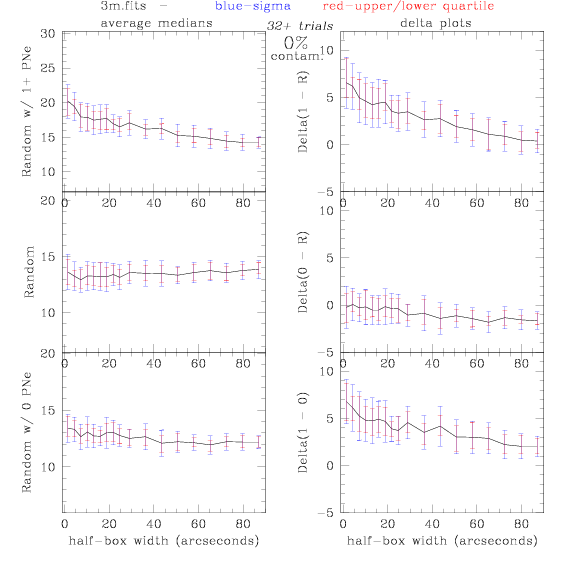

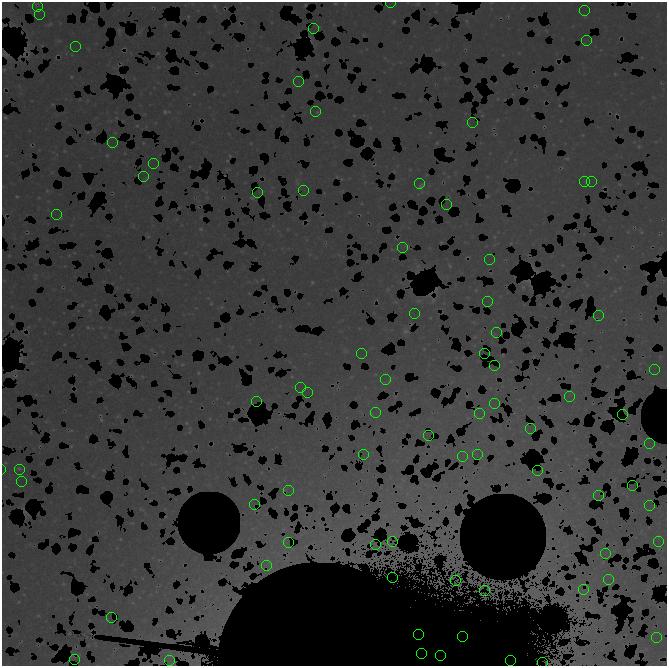
<-- less
contamination more contamination -->
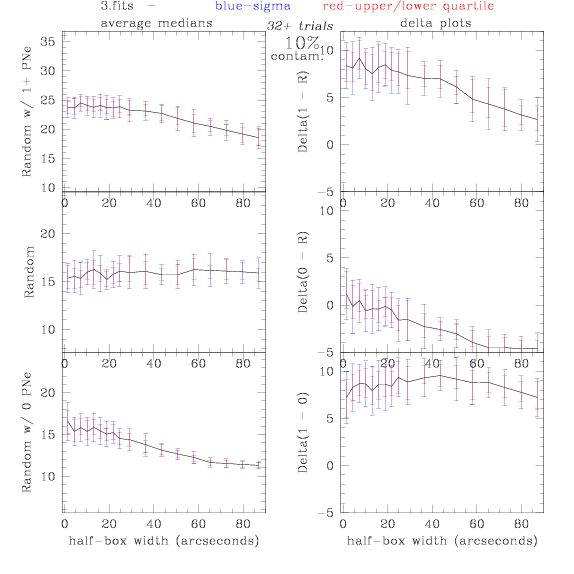
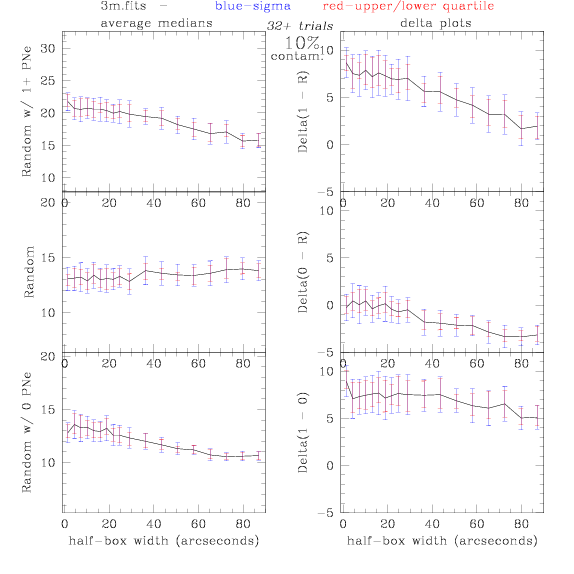

<-- less
contamination more contamination -->

<-- less
contamination more contamination -->

<-- less
contamination more contamination -->
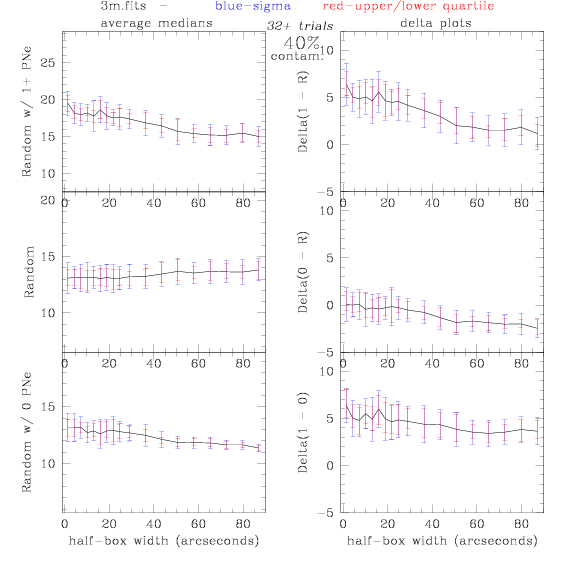

<-- less
contamination more contamination -->
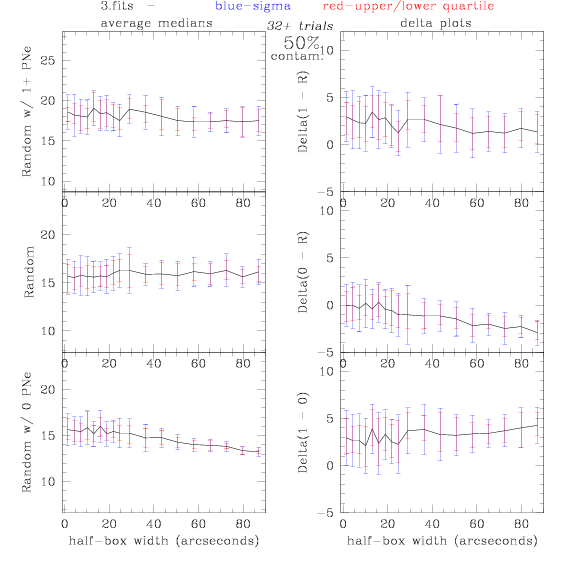
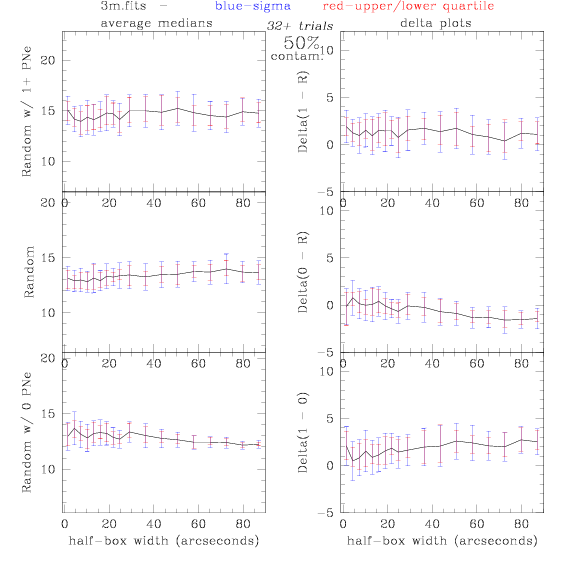

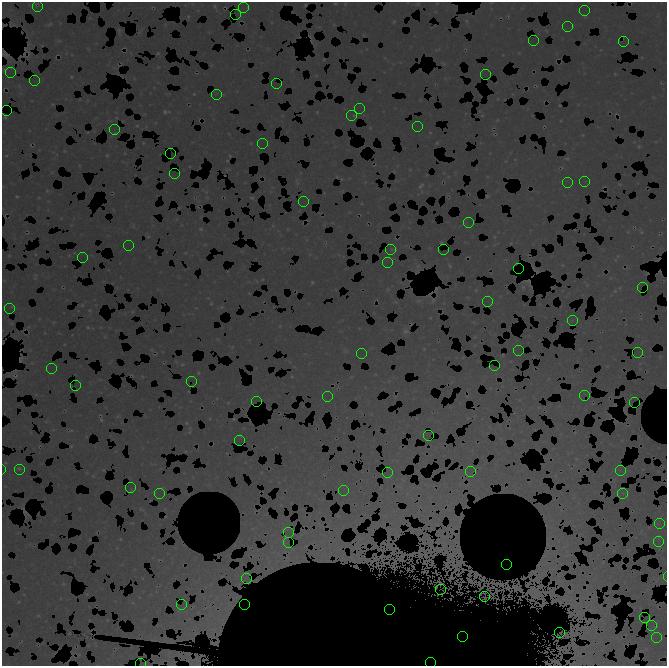
<-- less
contamination more contamination -->
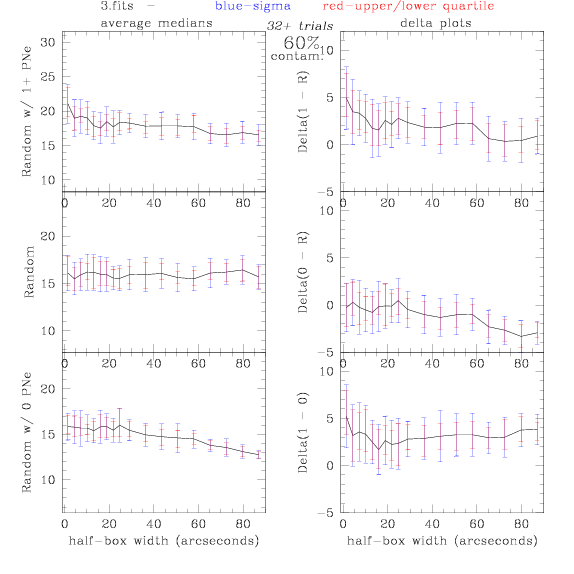
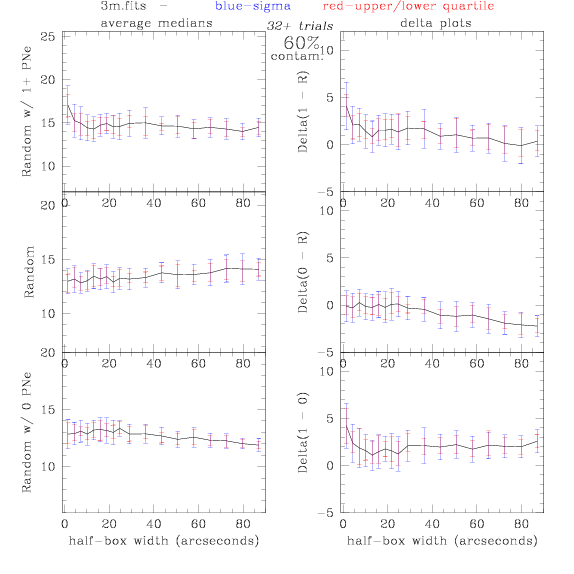

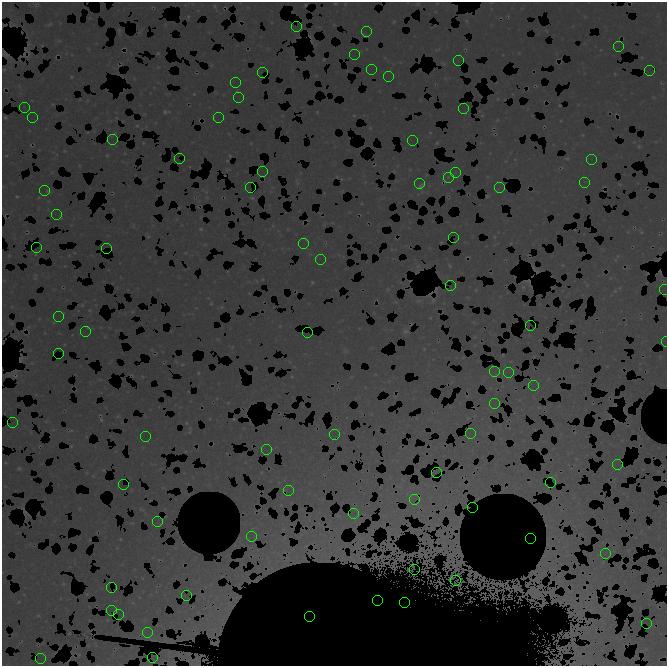
<-- less
contamination more contamination -->
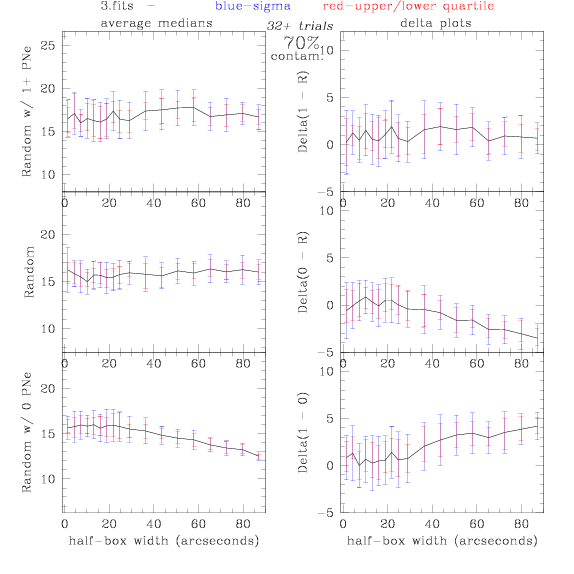
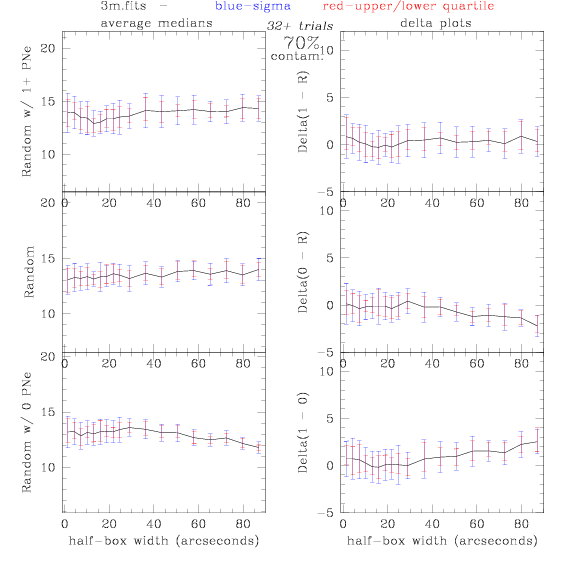

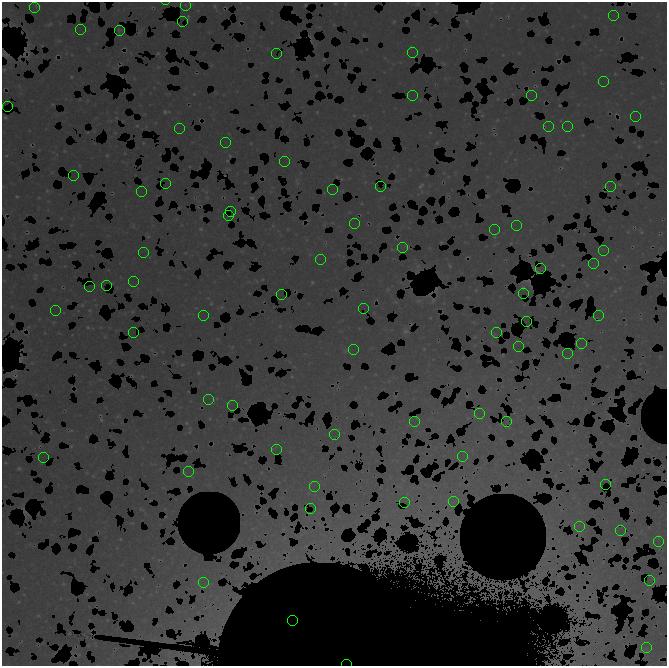
<-- less
contamination more contamination -->
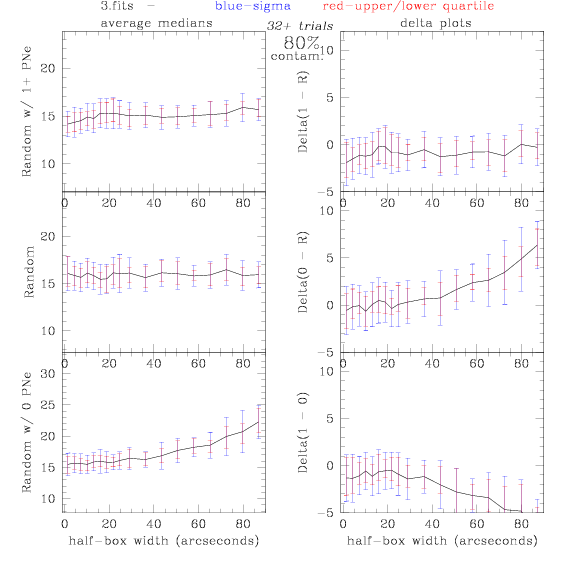
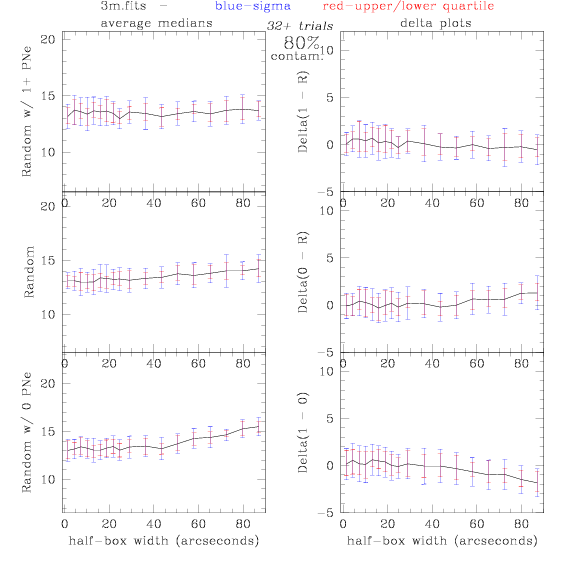

<-- less
contamination more contamination -->
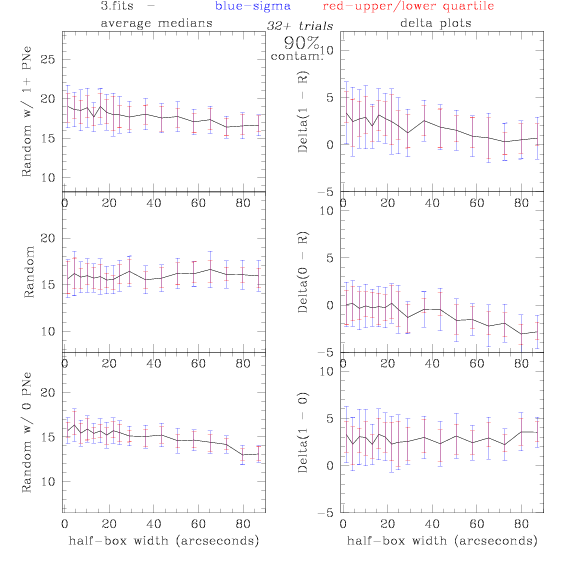
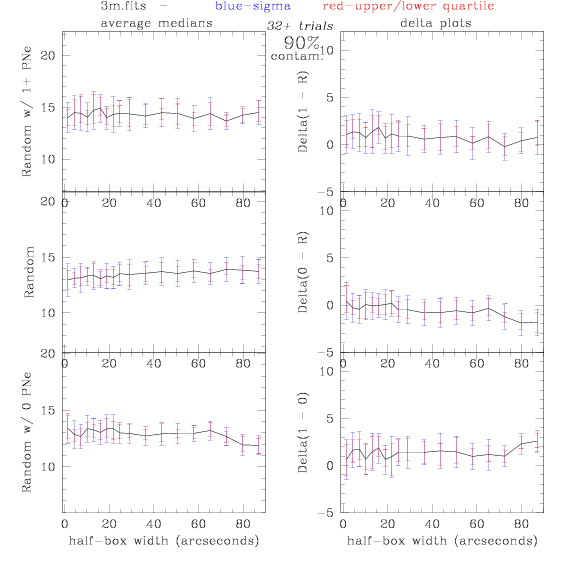

<-- less
contamination more contamination -->
back to 0% contamination
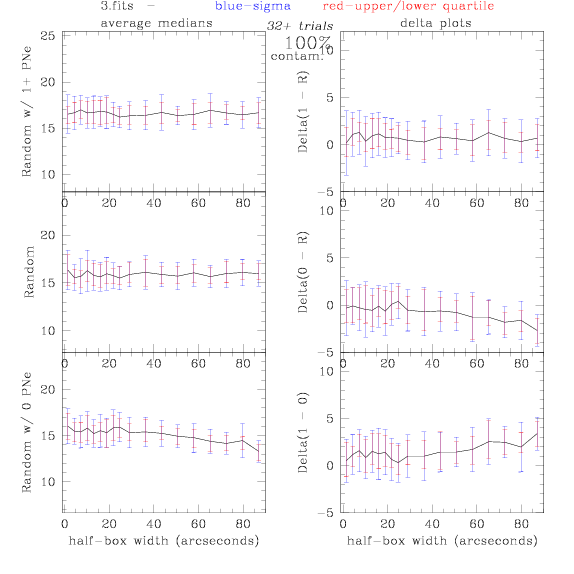
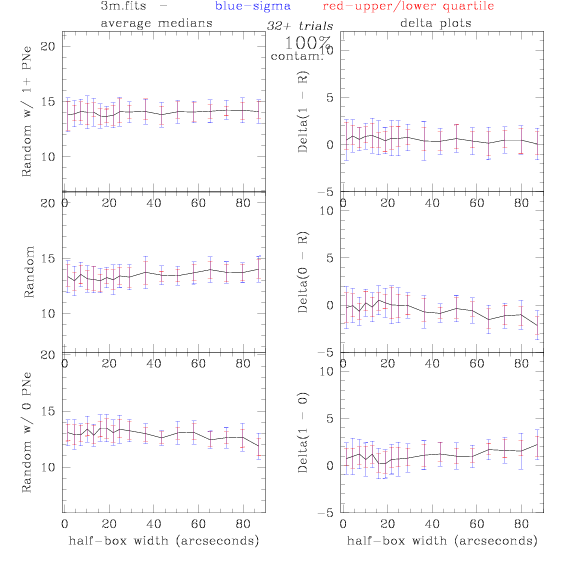
4.fits/4m.fits

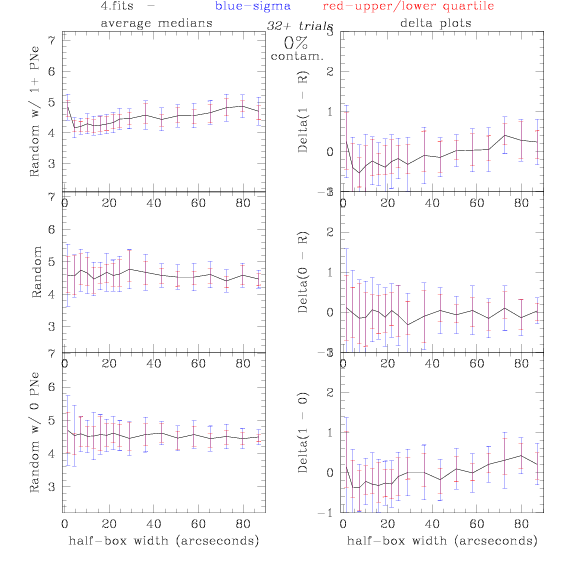
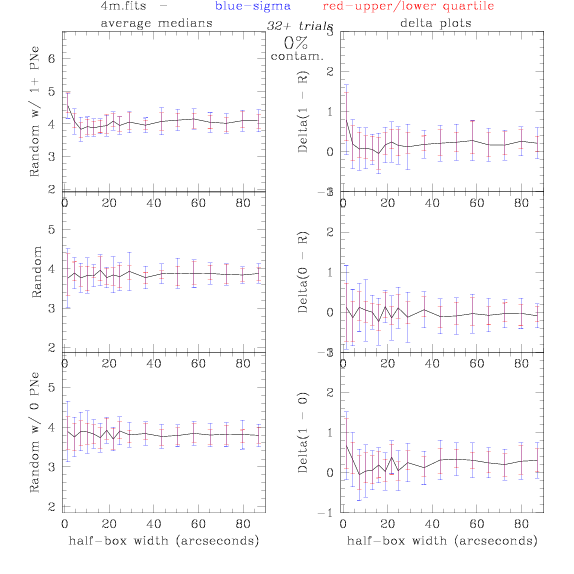

<-- less
contamination more contamination -->
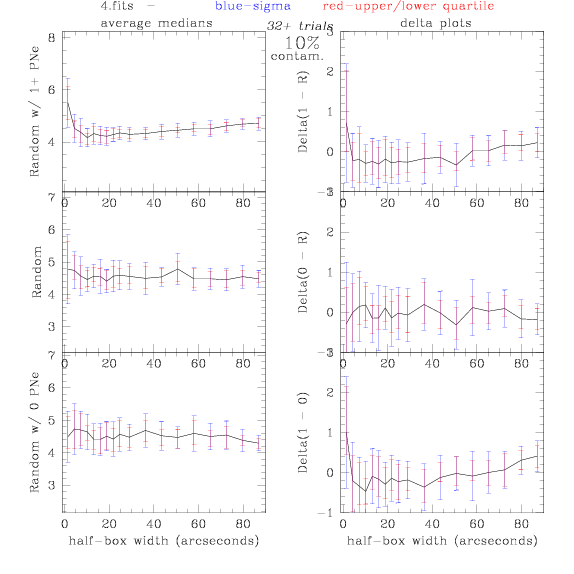
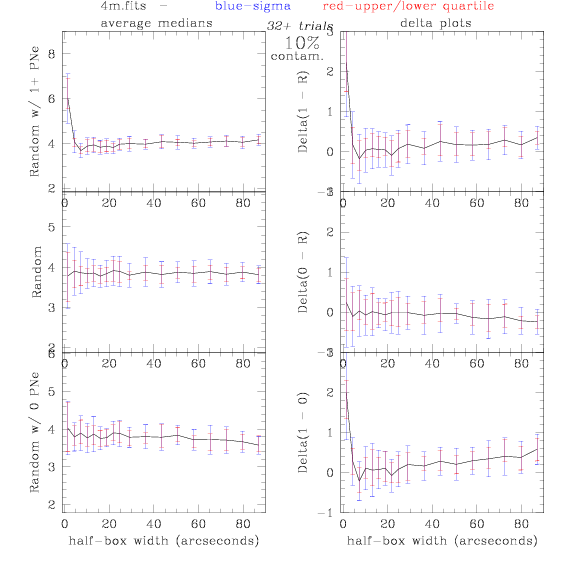

<-- less
contamination more contamination -->
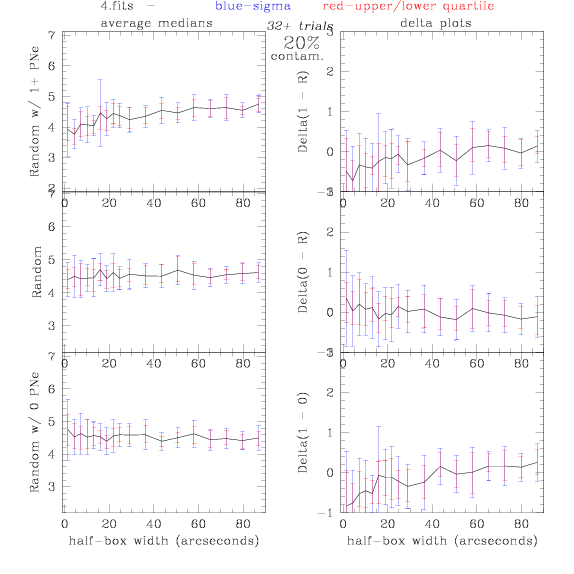
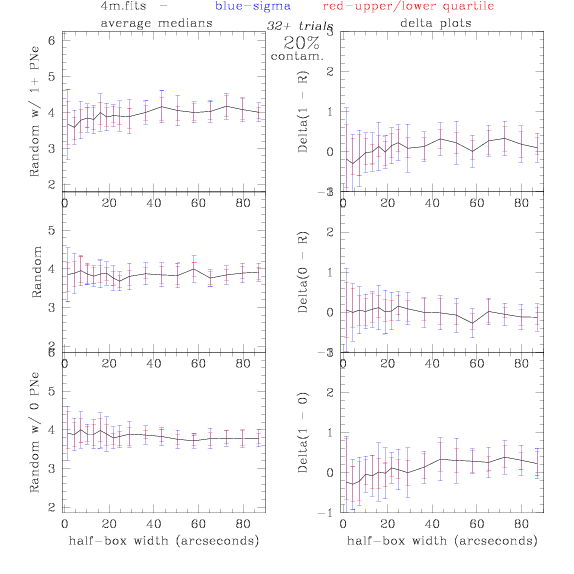

<-- less
contamination more contamination -->
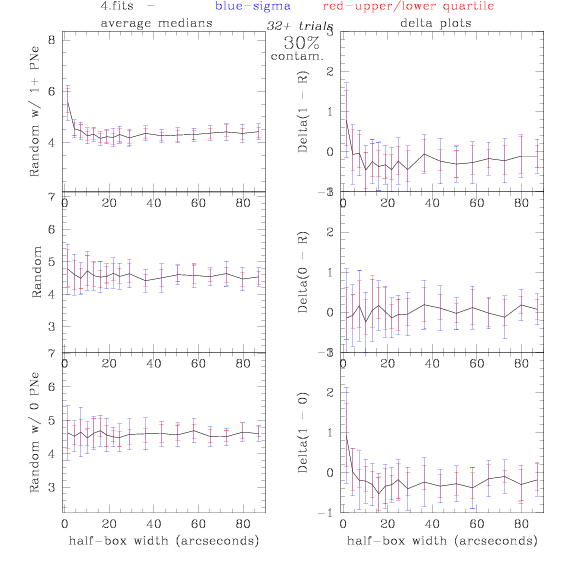
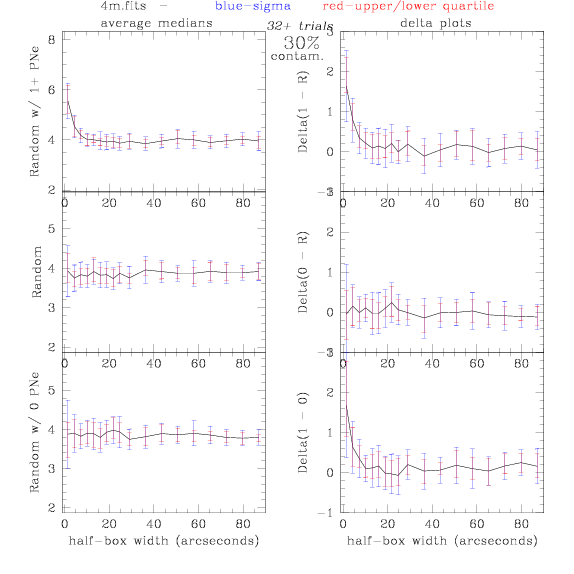

<-- less
contamination more contamination -->
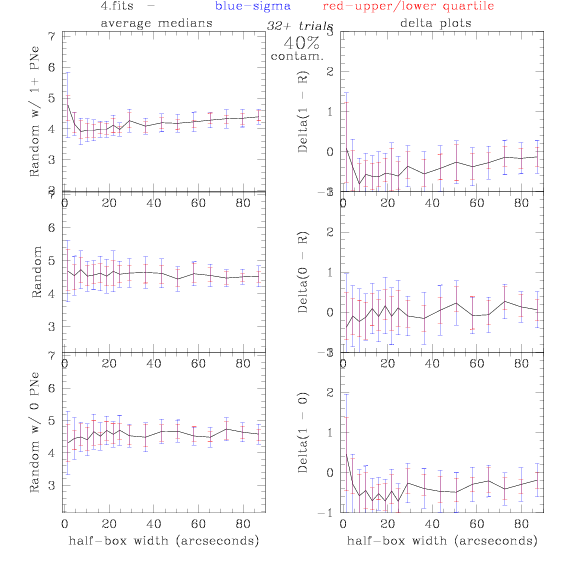
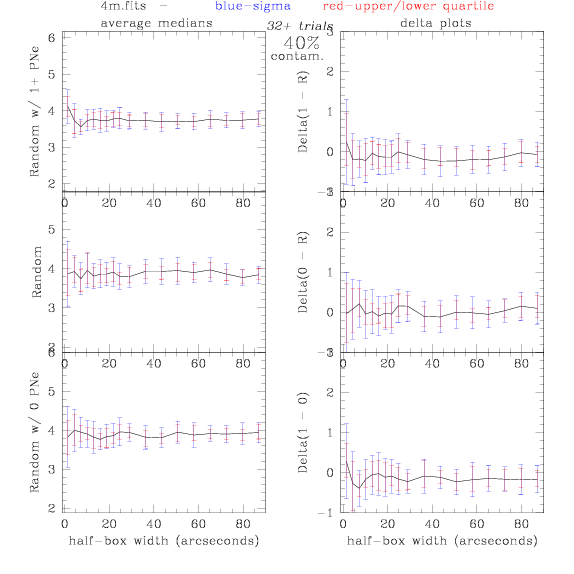

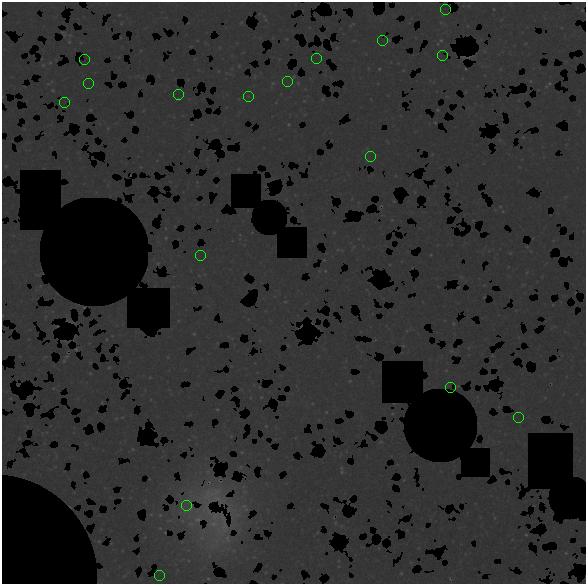
<-- less
contamination more contamination -->
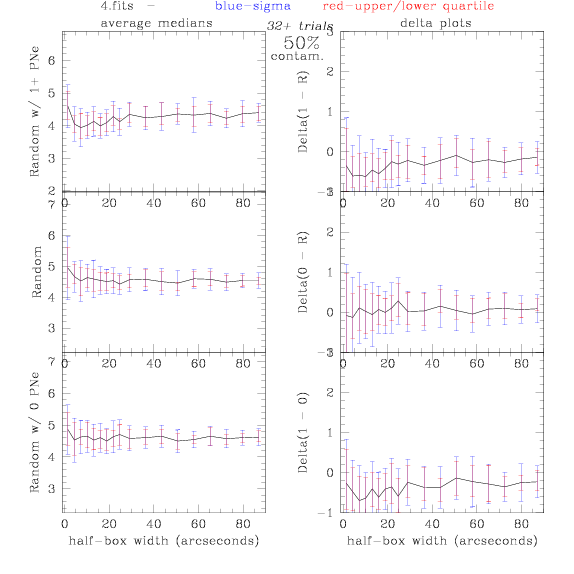
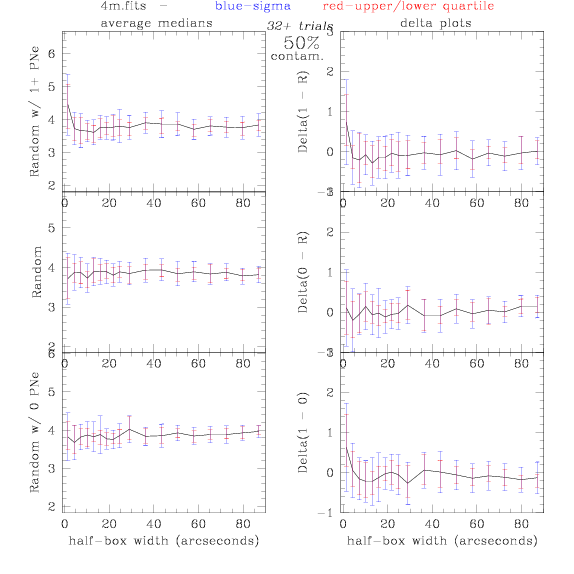

<-- less
contamination more contamination -->
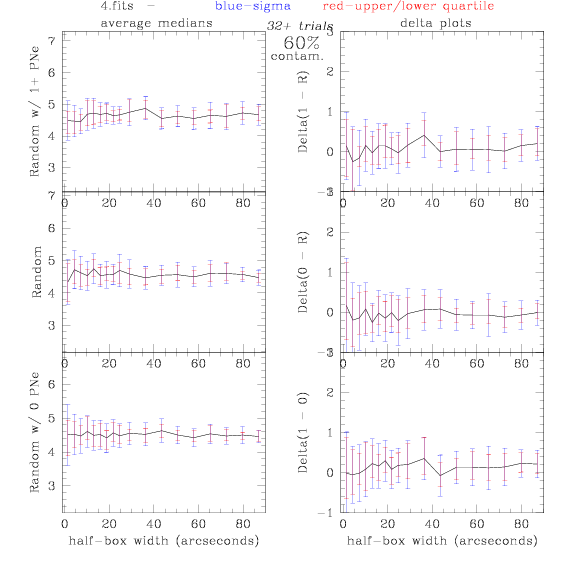
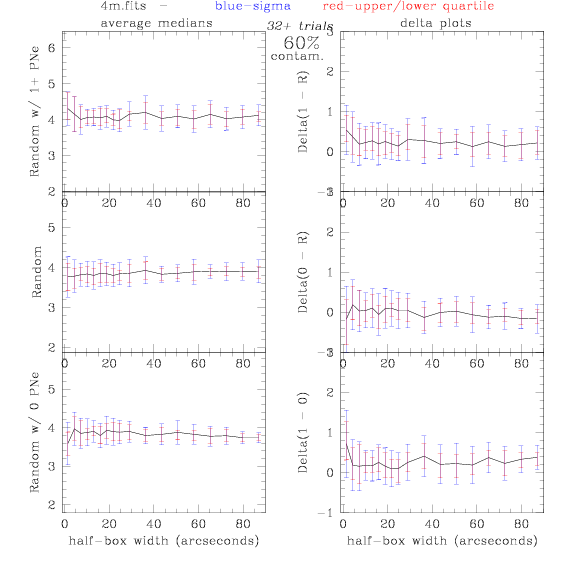

<-- less
contamination more contamination -->
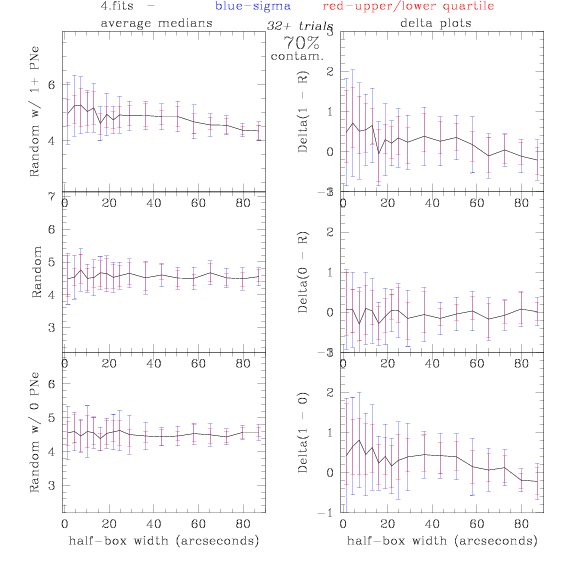
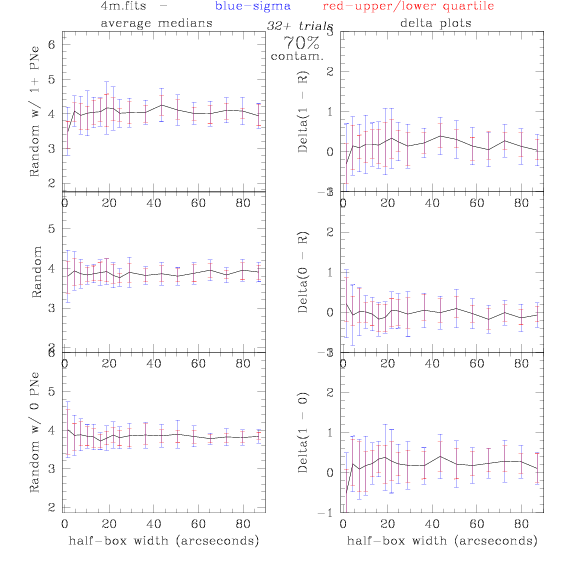

<-- less
contamination more contamination -->
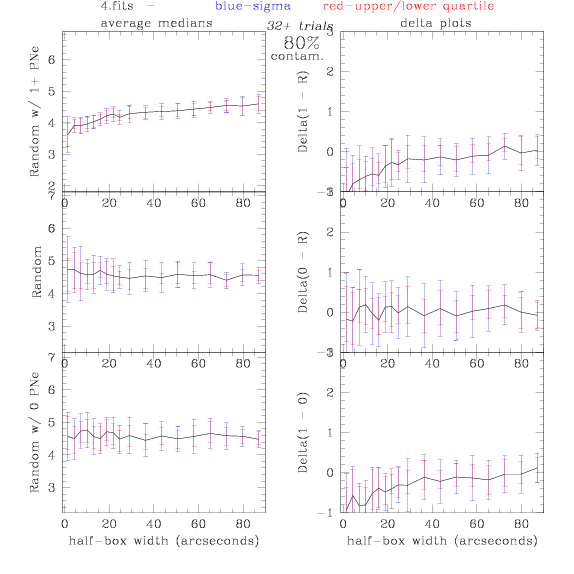
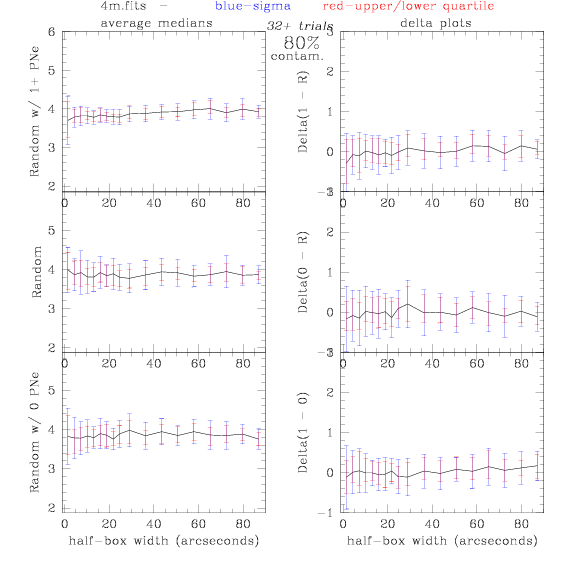

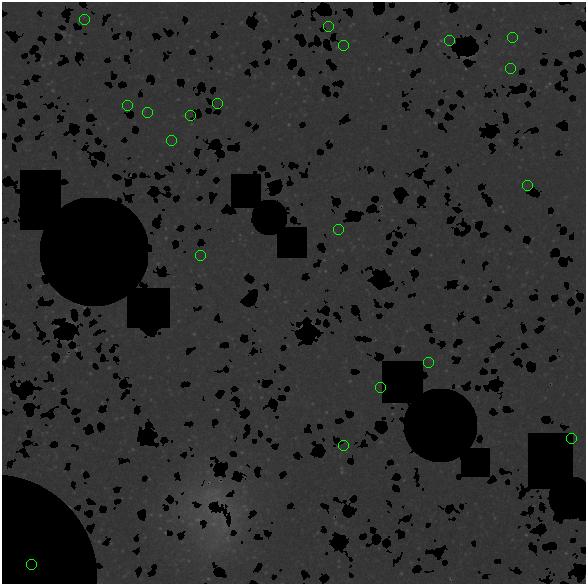
<-- less
contamination more contamination -->
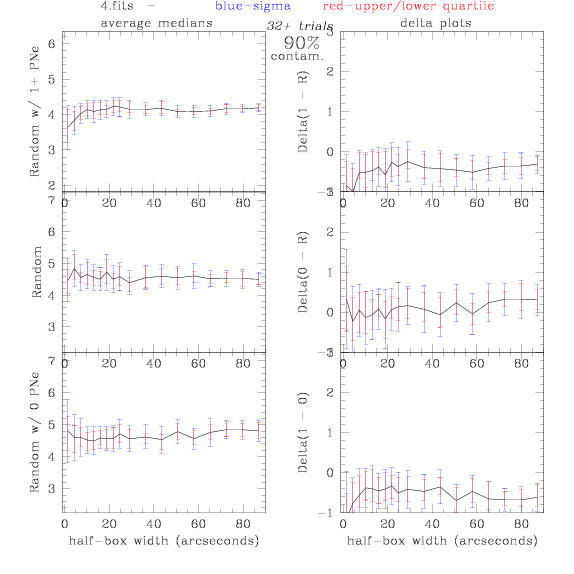
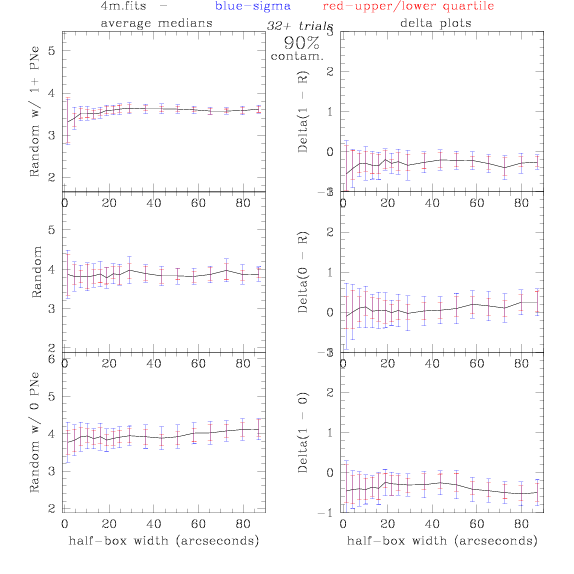

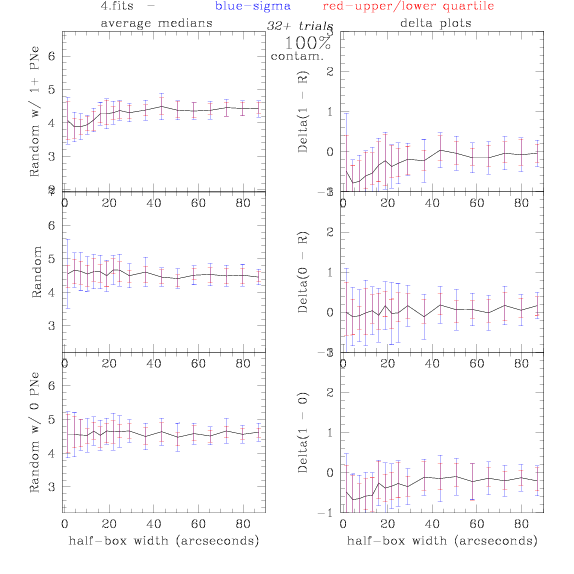
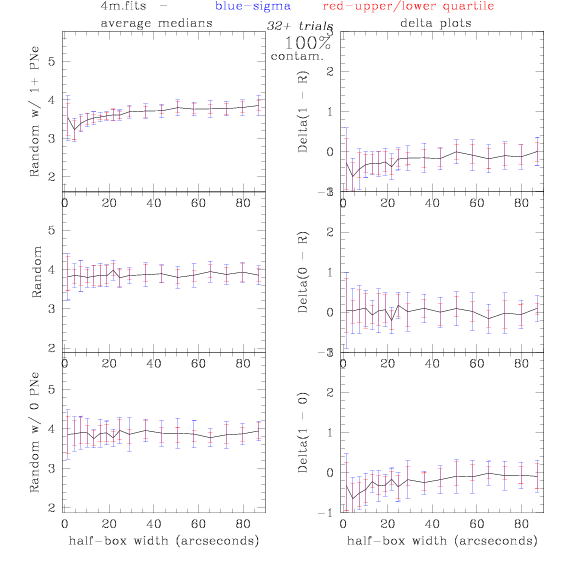
<-- less
contamination more contamination -->
back to 0% contamination
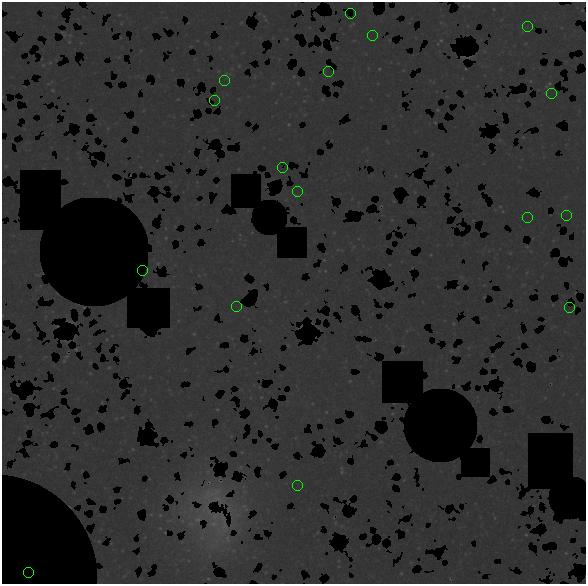
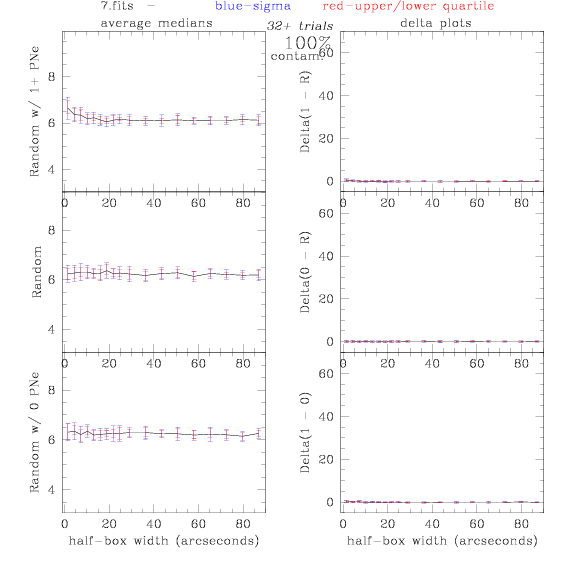
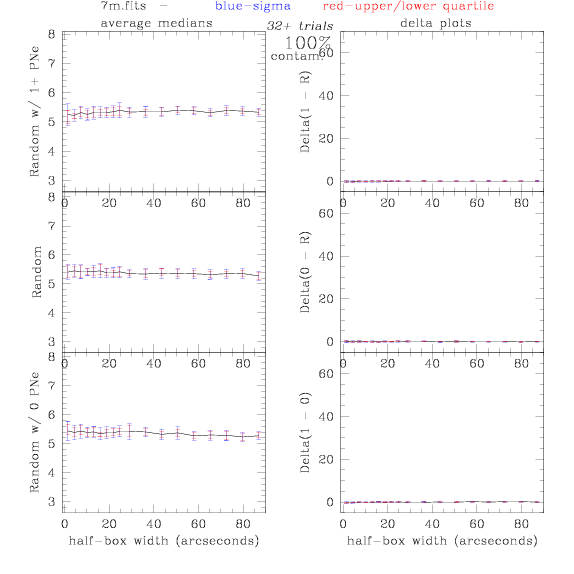
7.fits/7m.fits
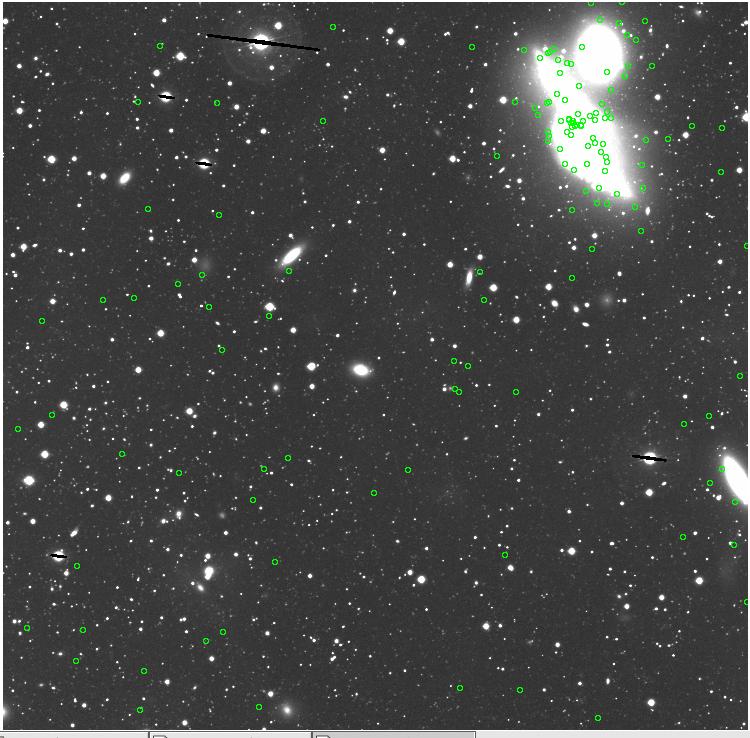
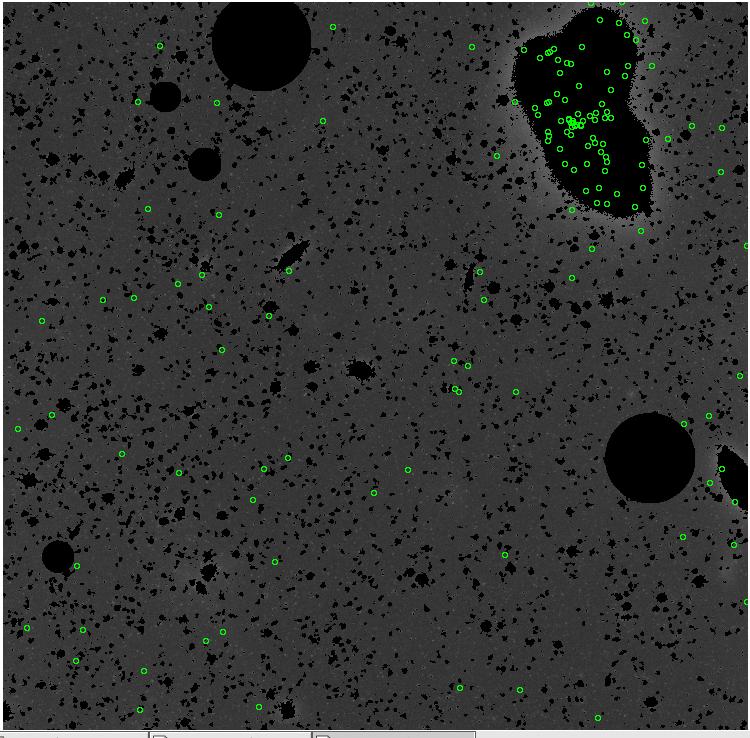
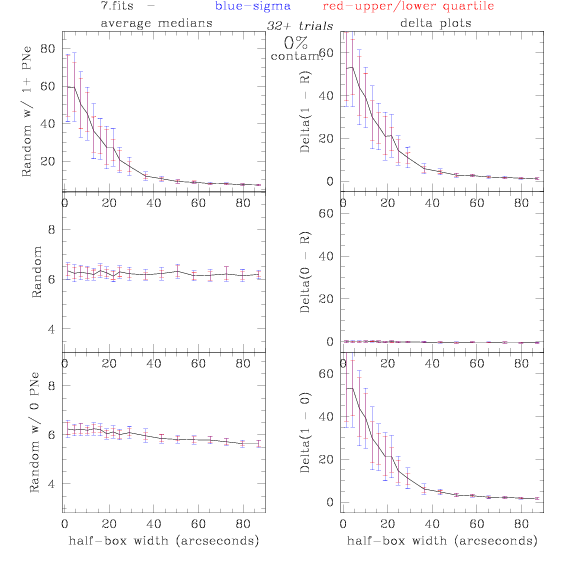
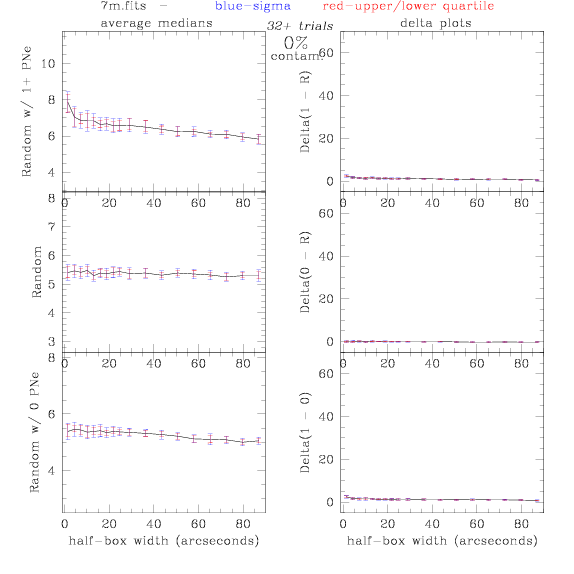
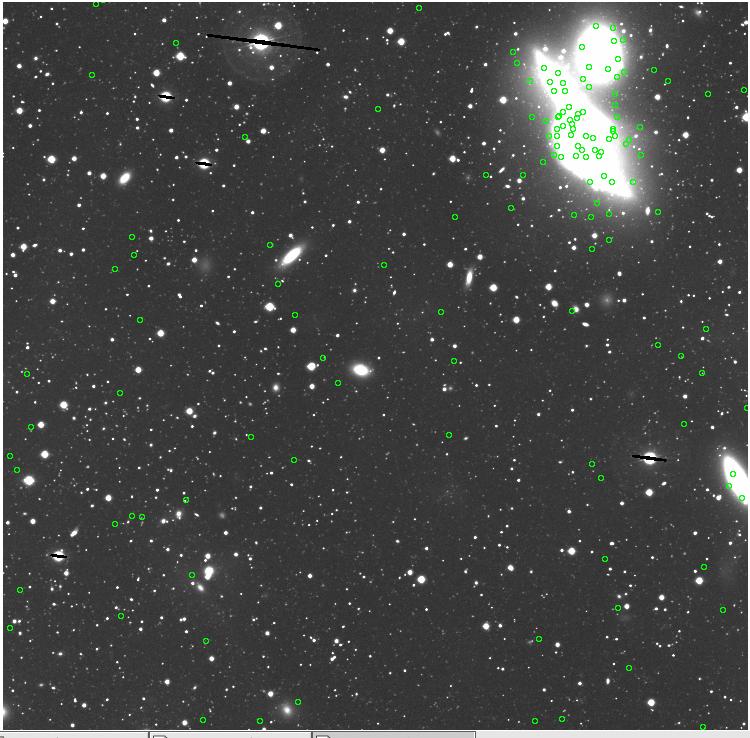
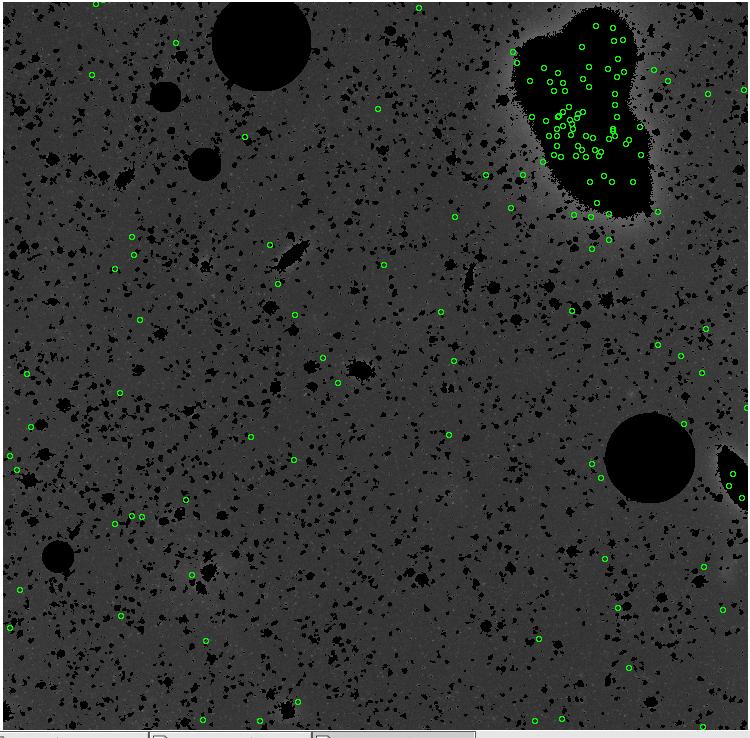
<-- less
contamination more contamination -->
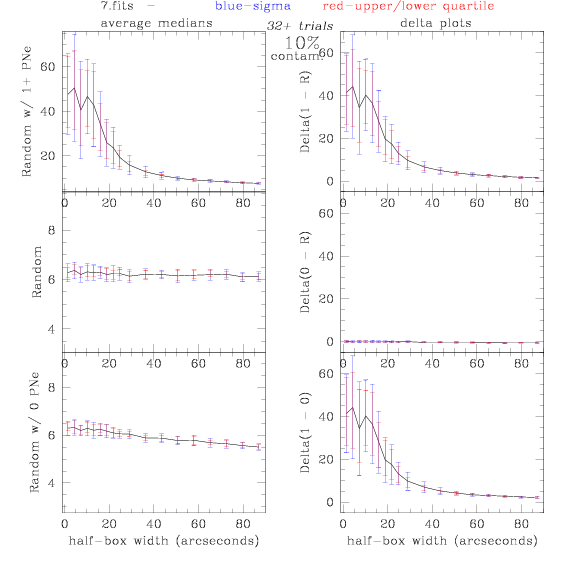
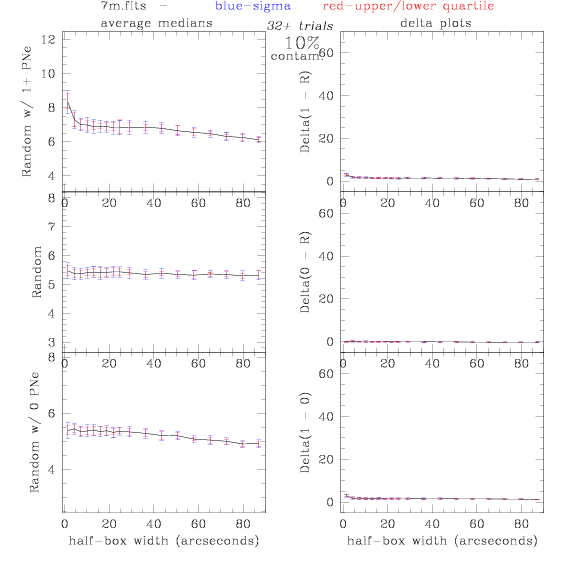

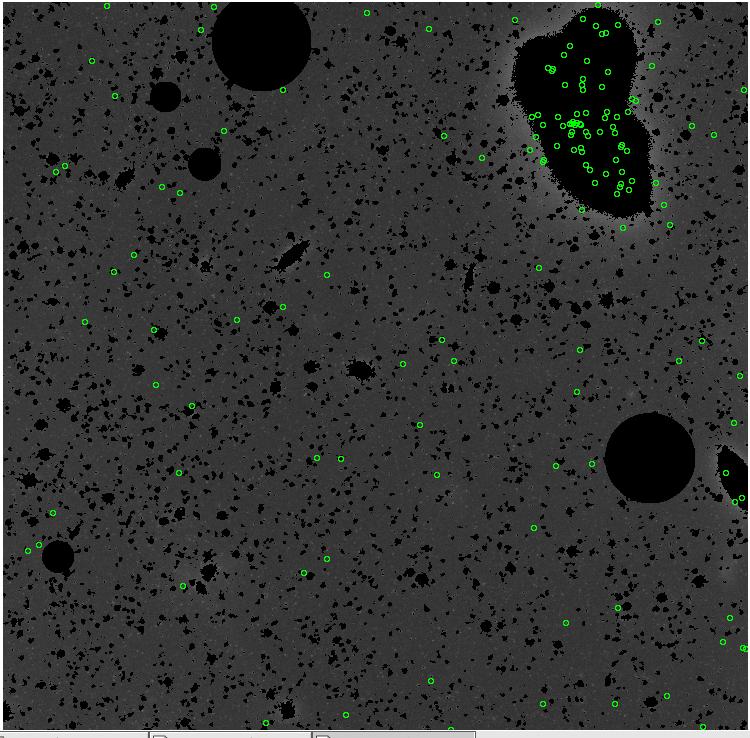
<-- less
contamination more contamination -->
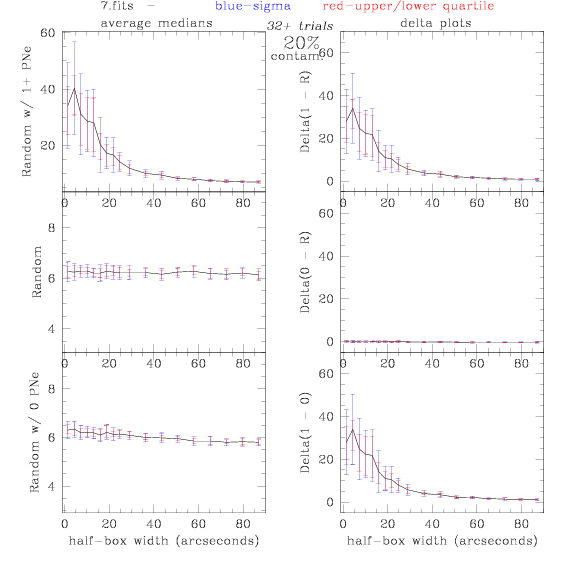
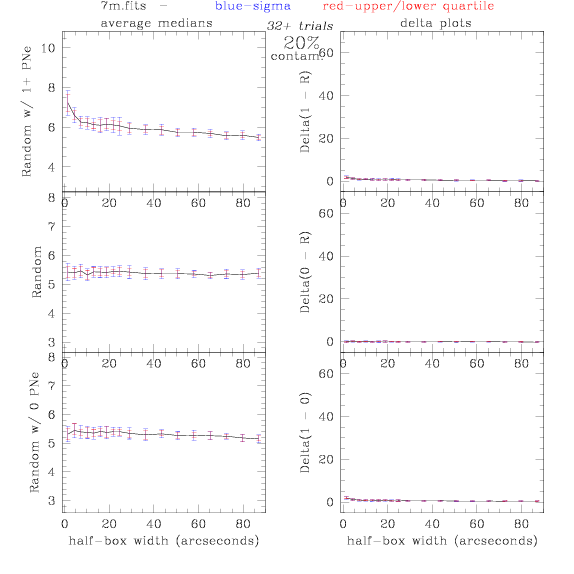

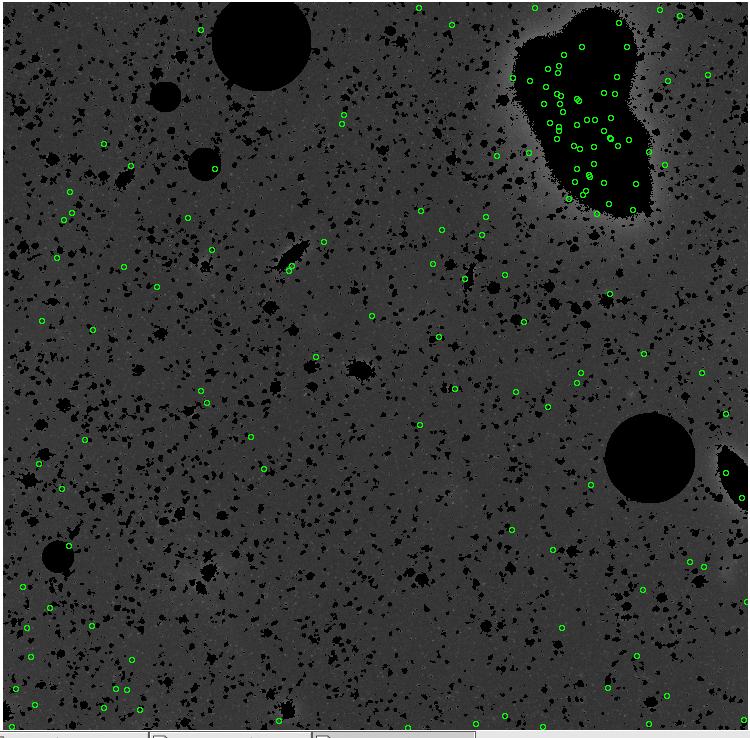
<-- less
contamination more contamination -->
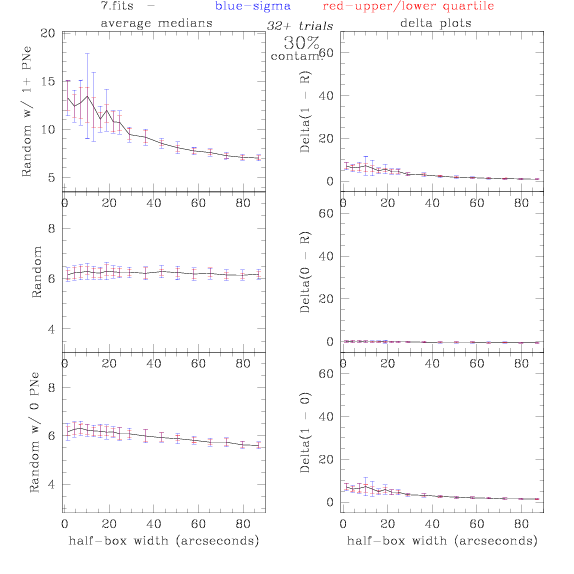
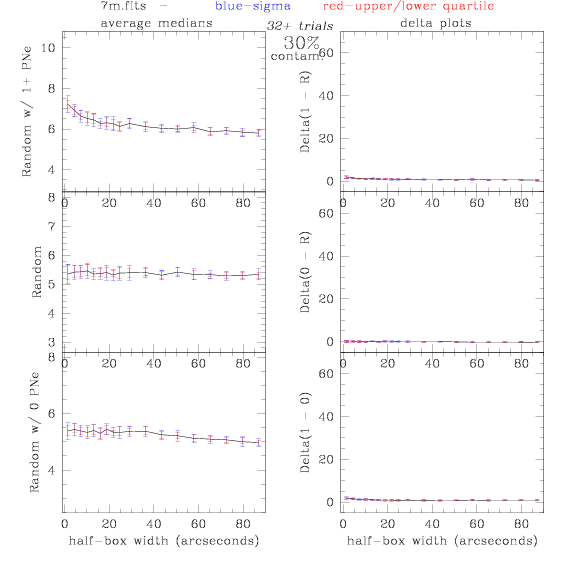
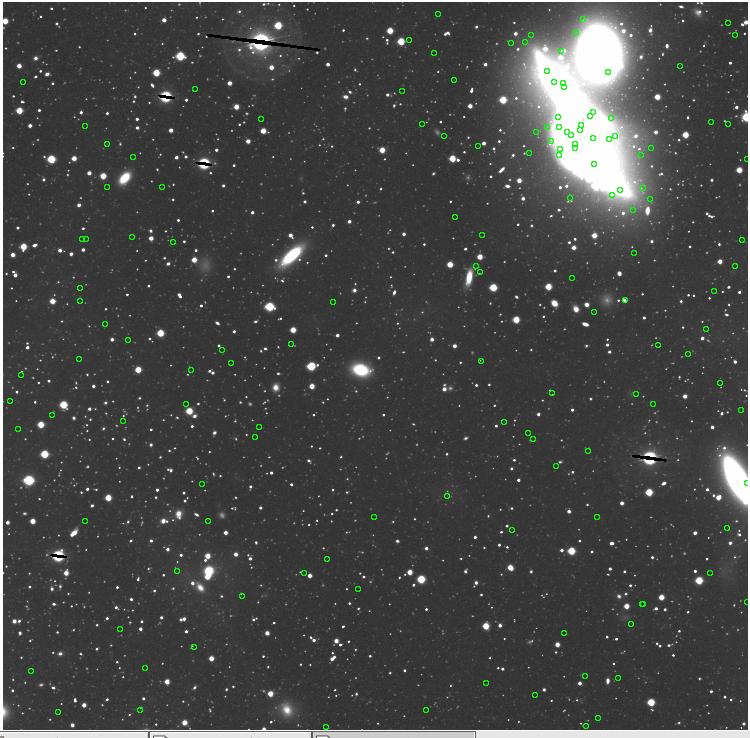
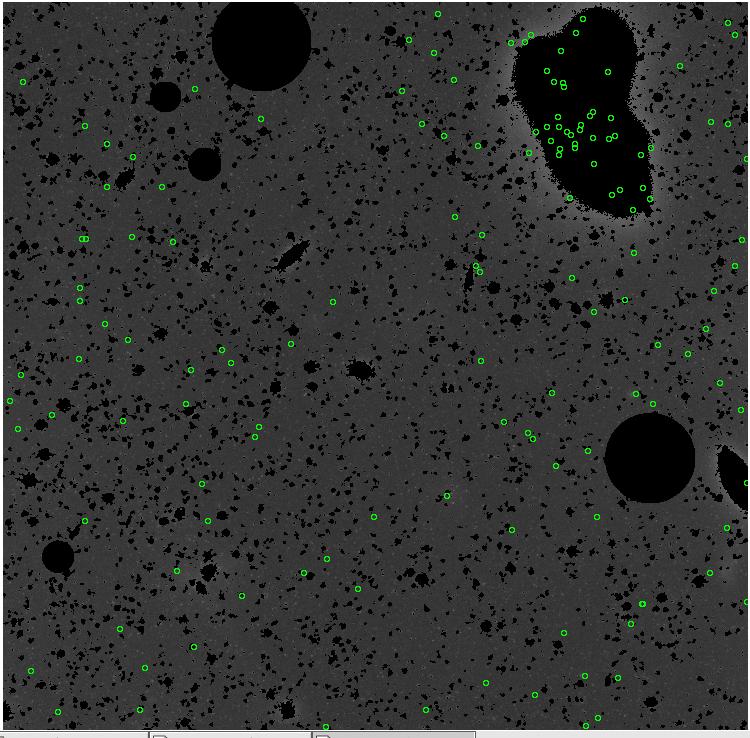
<-- less
contamination more contamination -->
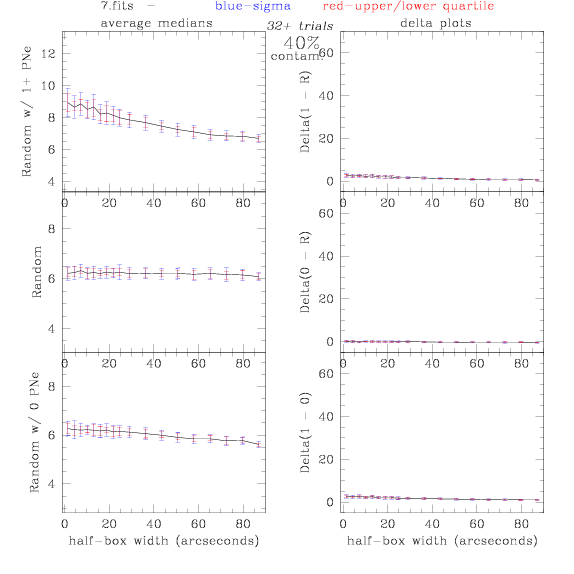
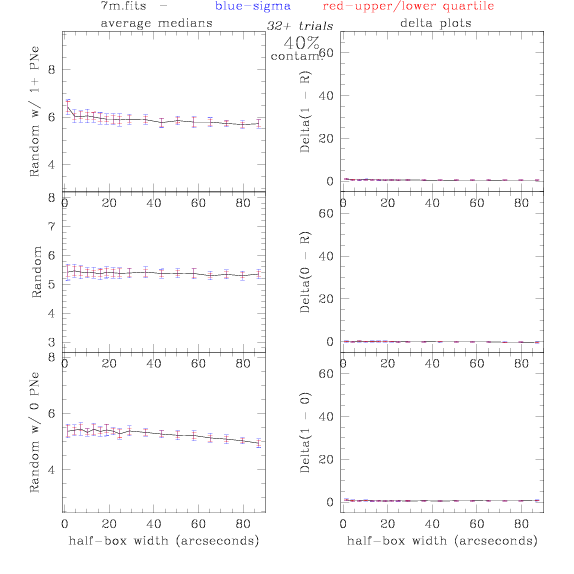
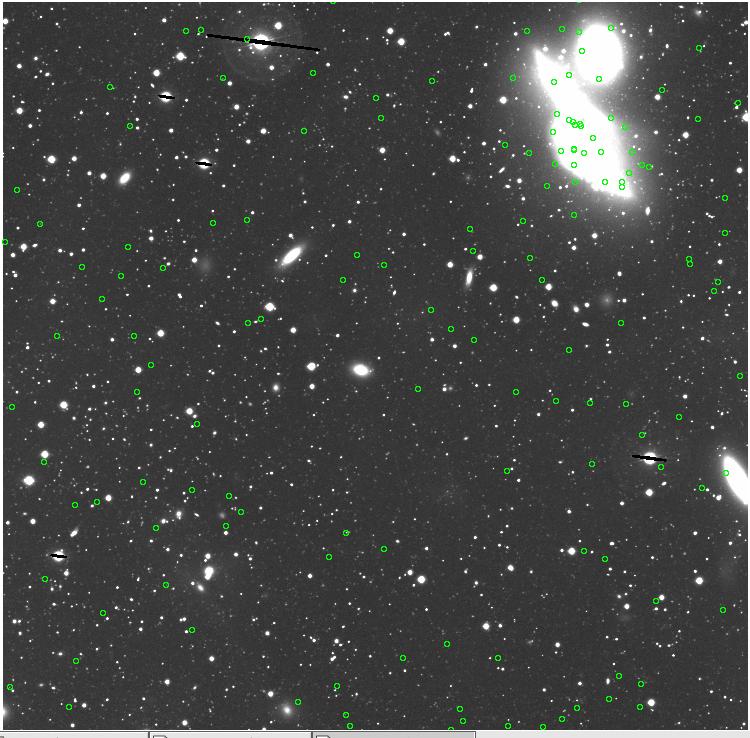
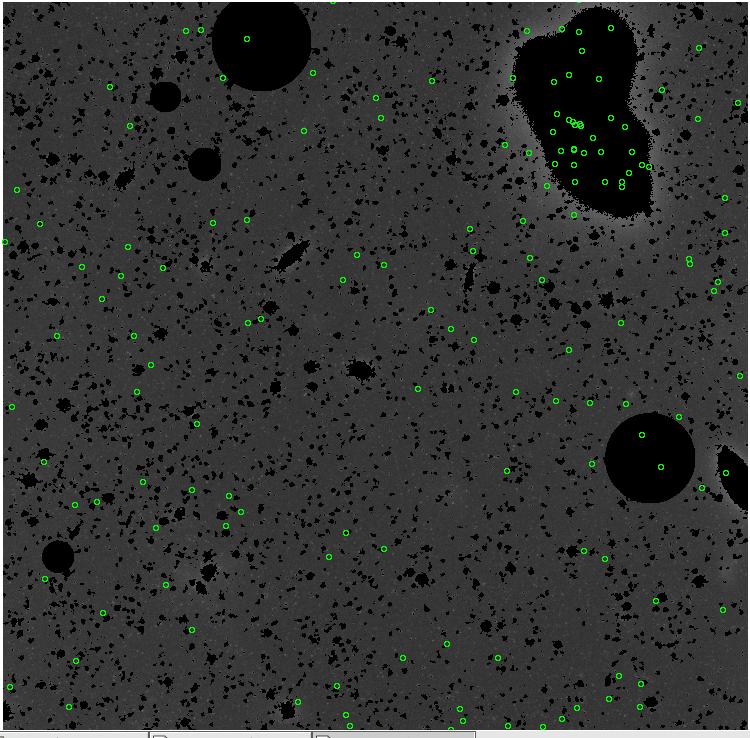
<-- less
contamination more contamination -->
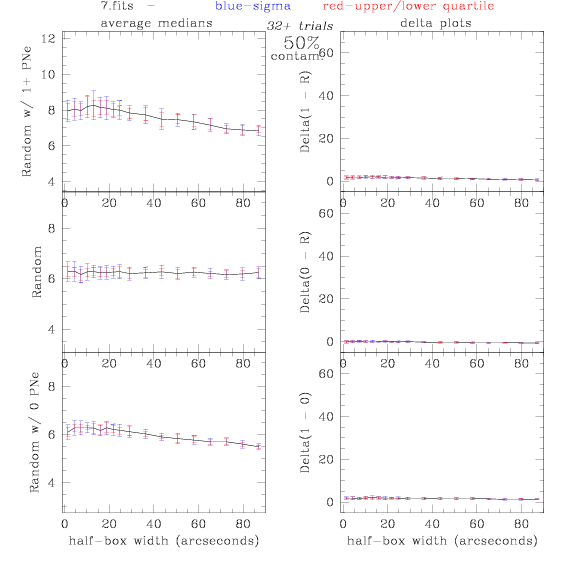
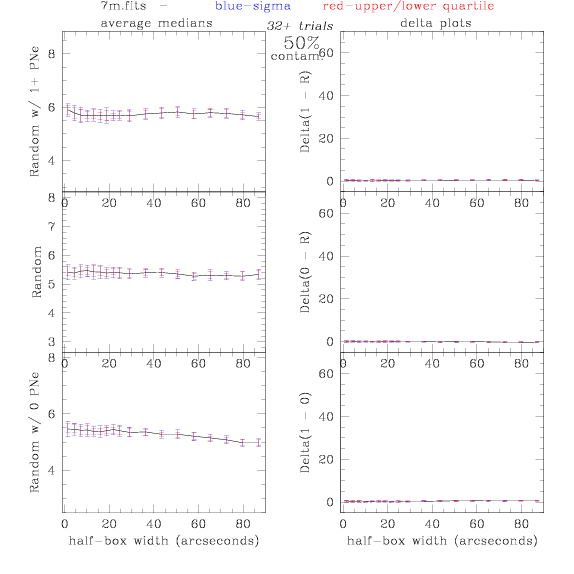

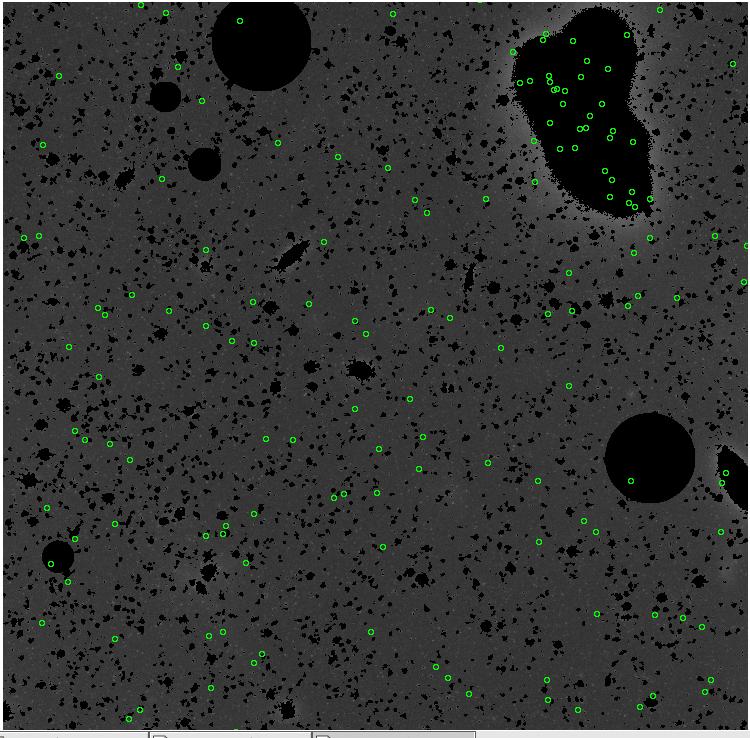
<-- less
contamination more contamination -->
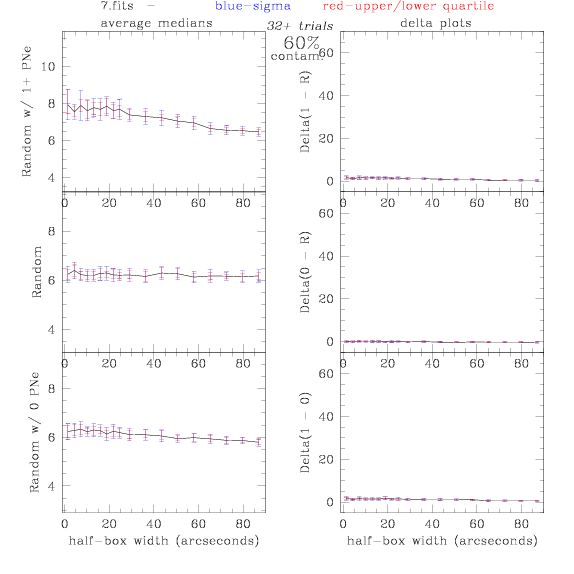
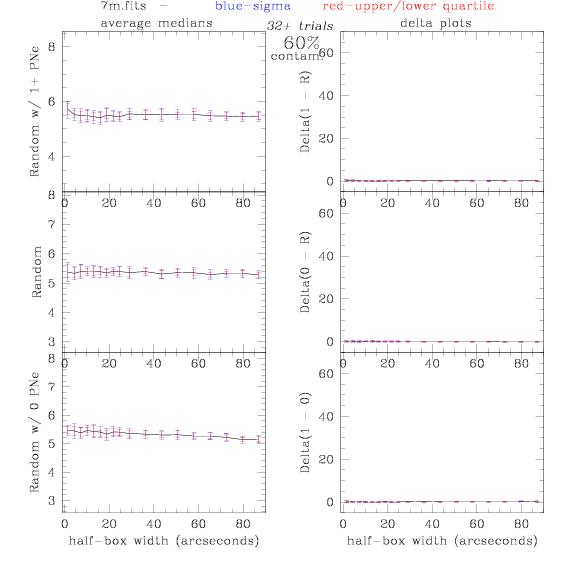
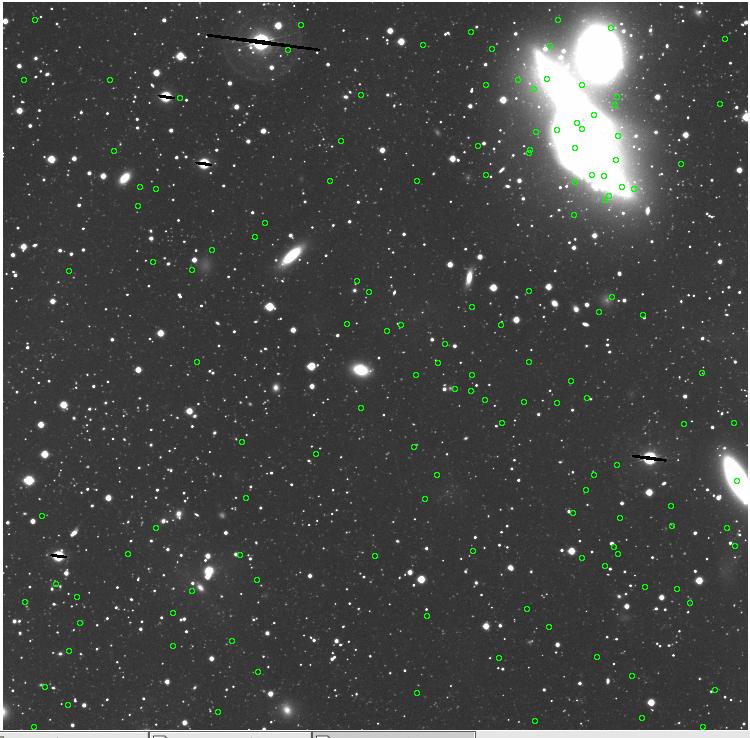
<-- less
contamination more contamination -->
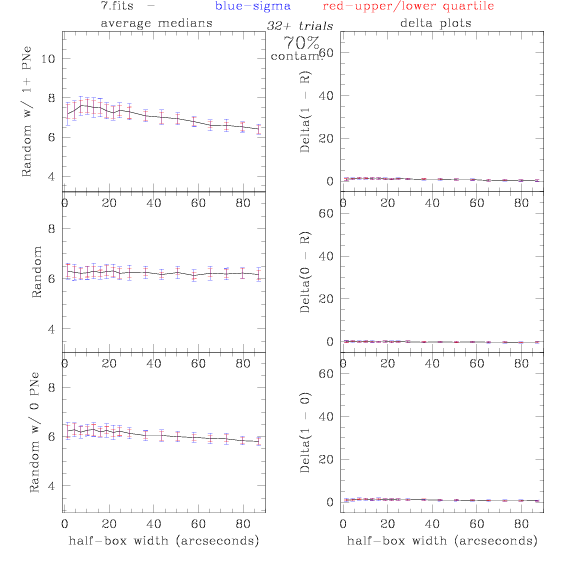
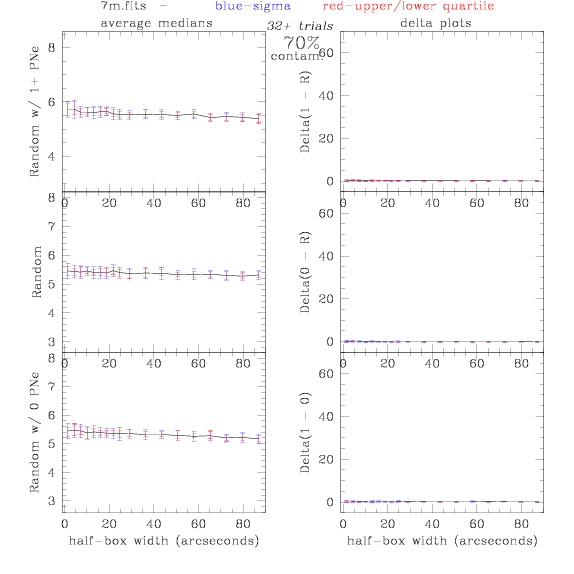

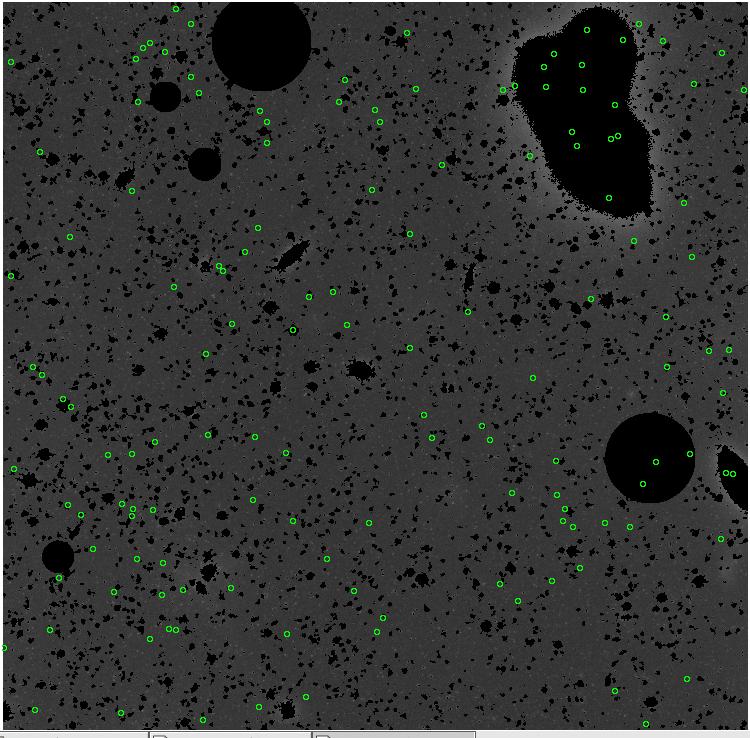
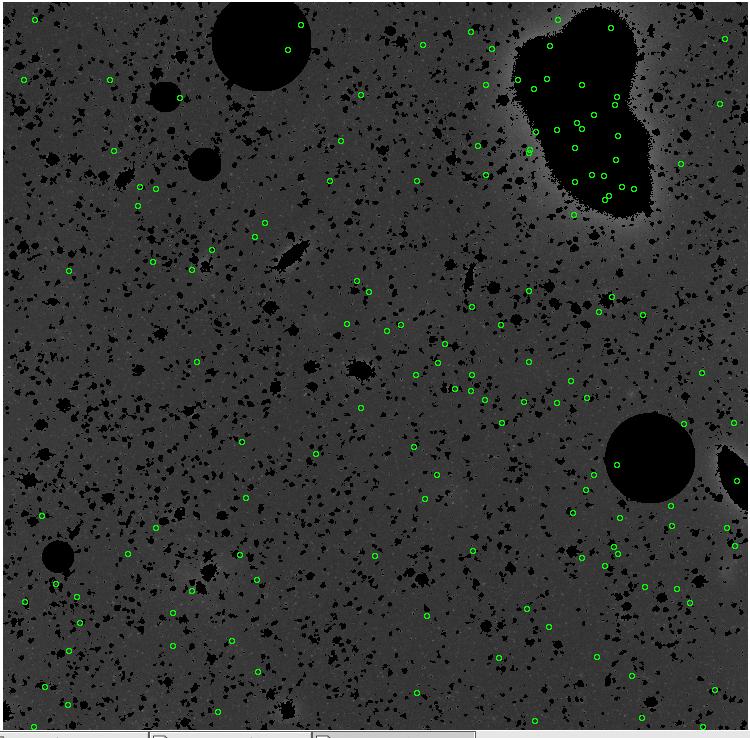
<-- less
contamination more contamination -->
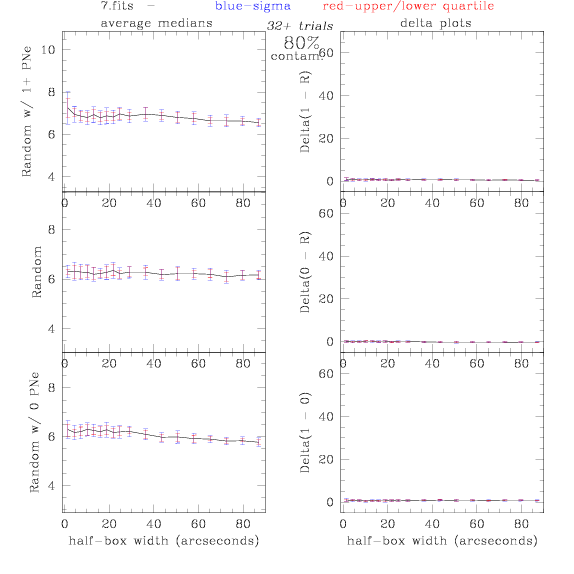
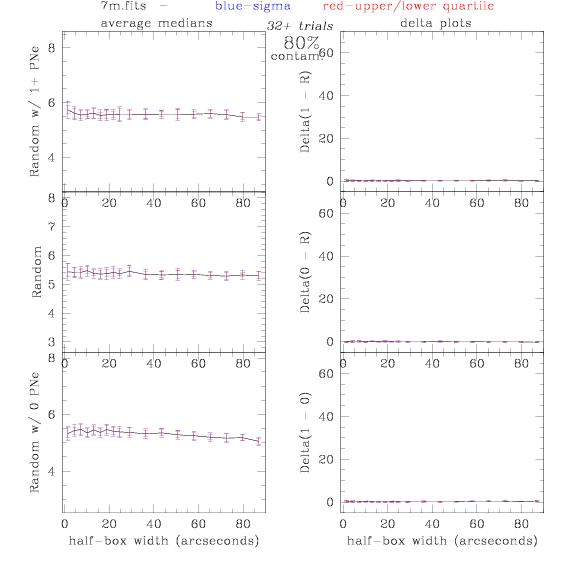

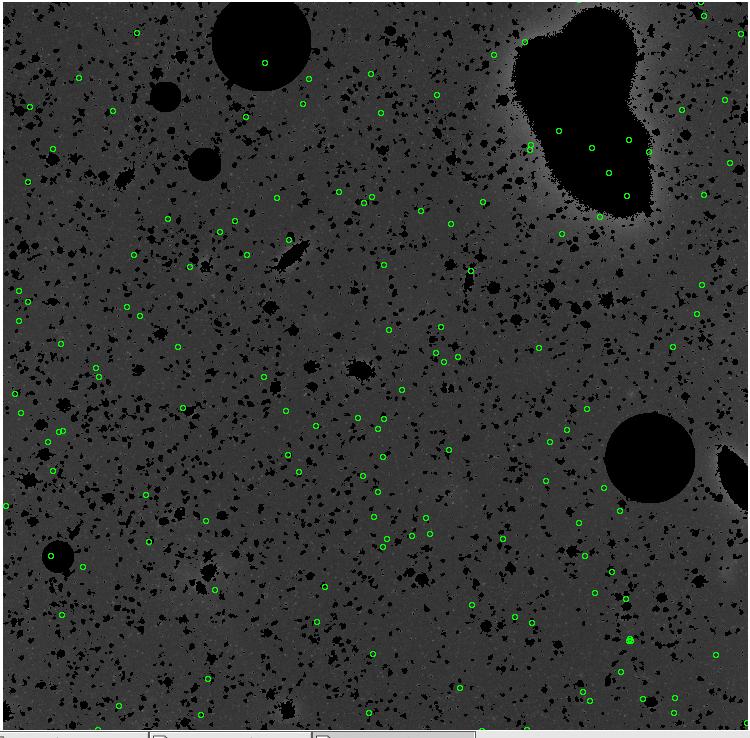
<-- less
contamination more contamination -->
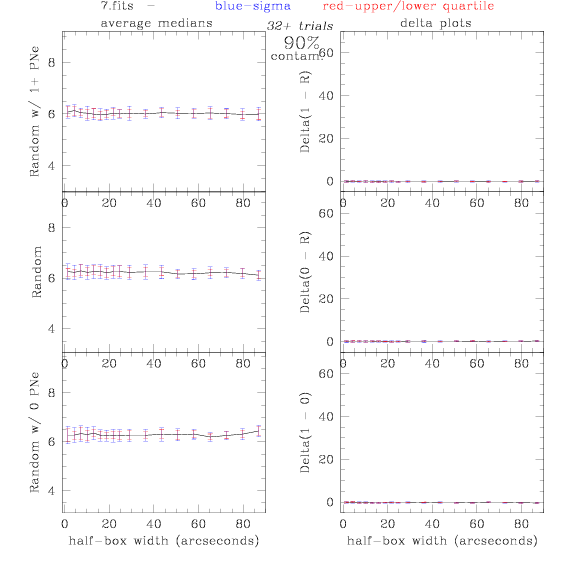
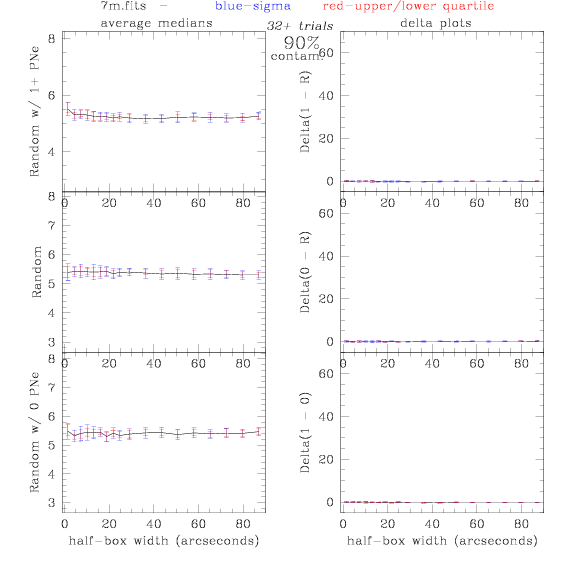
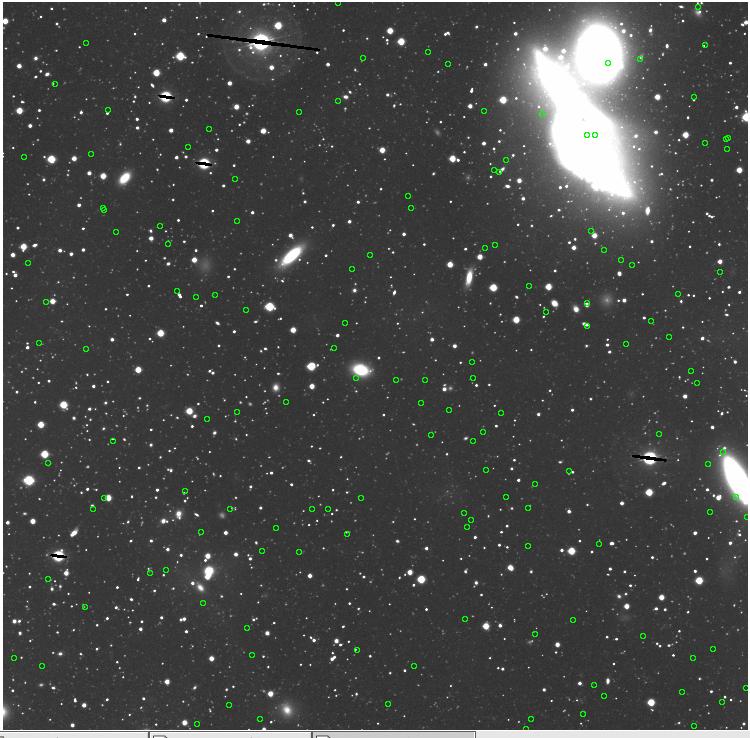
<-- less
contamination more contamination -->
back to 0% contamination
Sub.fits/Subm.fits

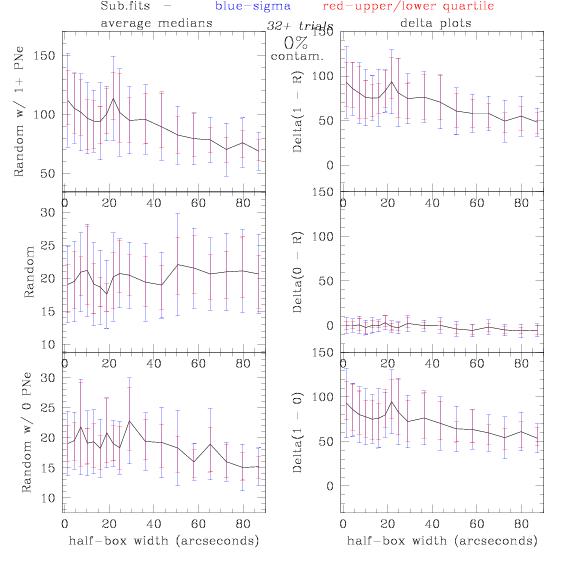
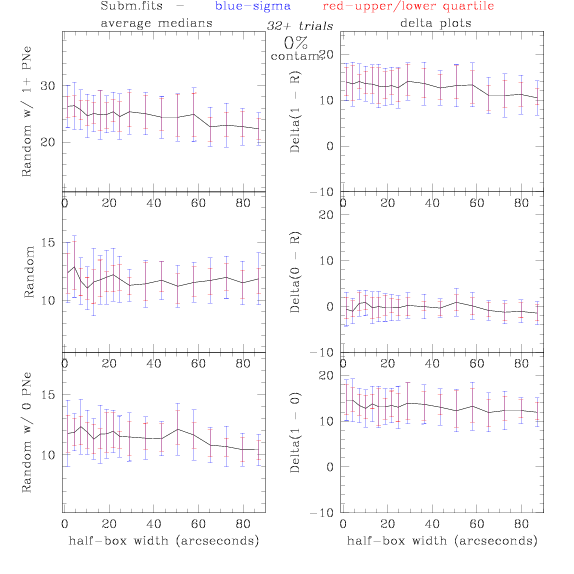

<-- less
contamination more contamination -->
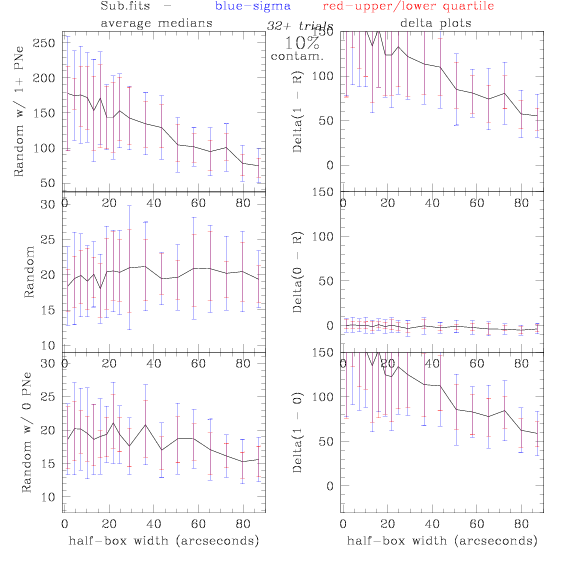
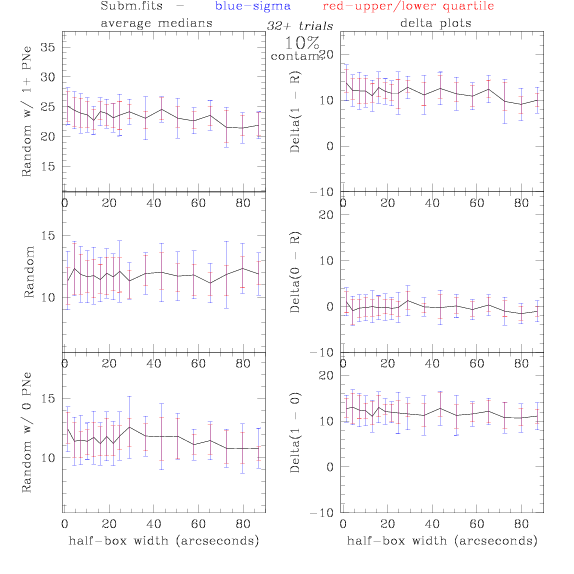

<-- less
contamination more contamination -->
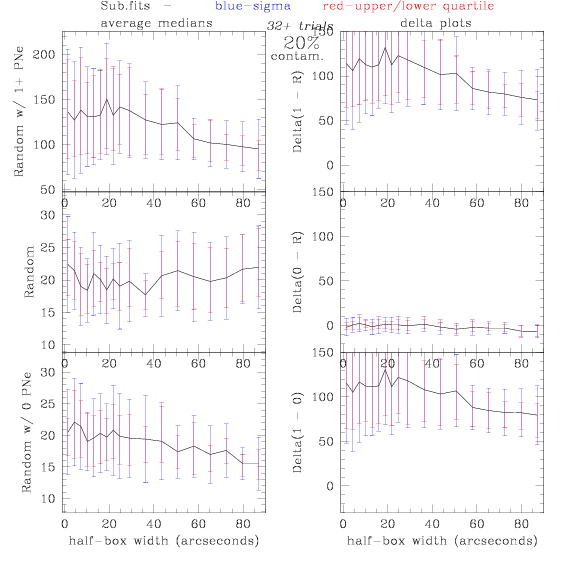
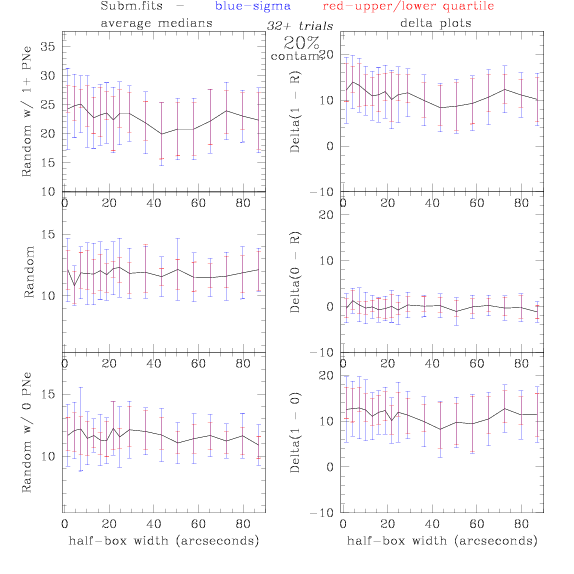

<-- less
contamination more contamination -->
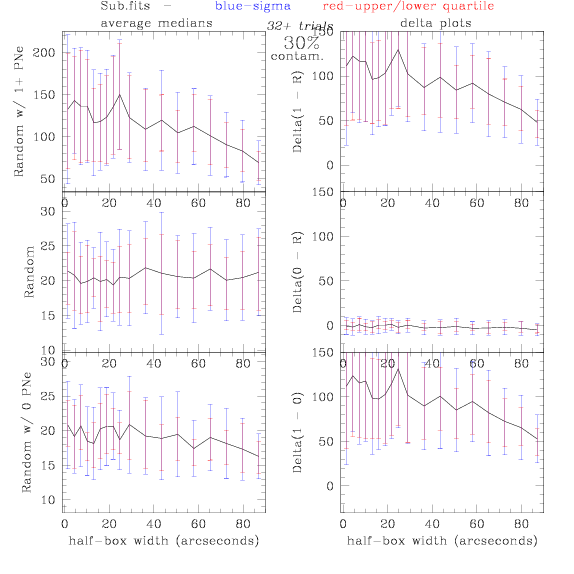
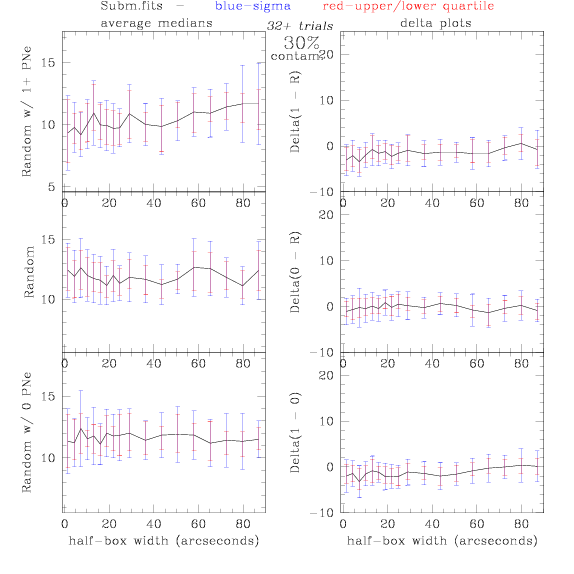

<-- less
contamination more contamination -->
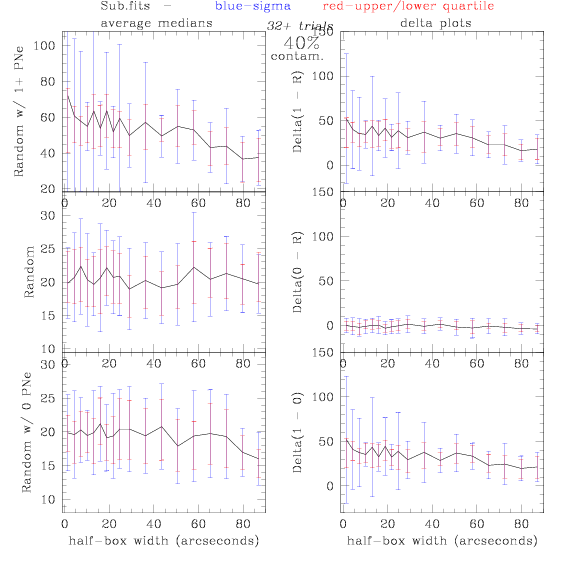
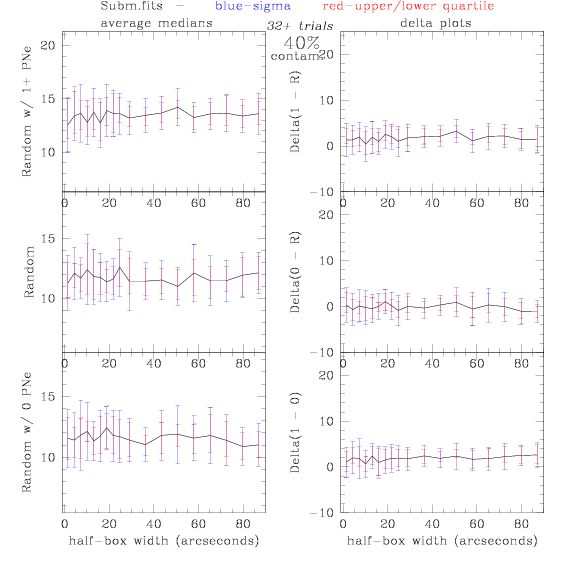

<-- less
contamination more contamination -->
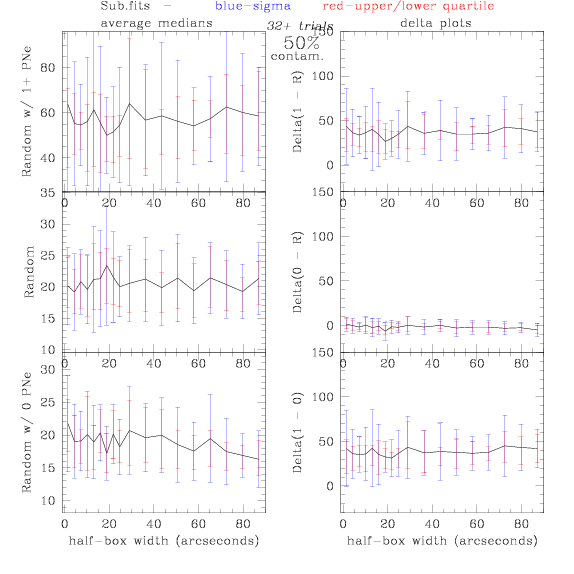
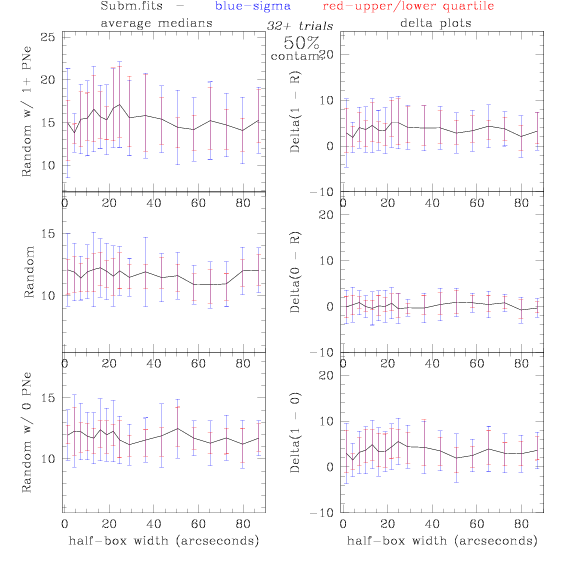

<-- less
contamination more contamination -->
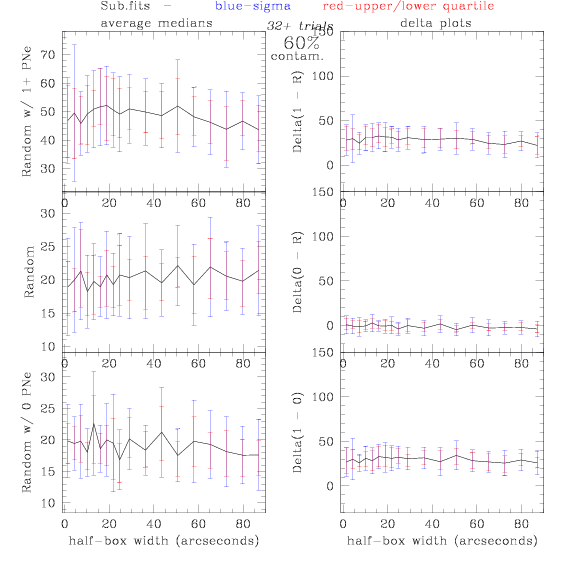
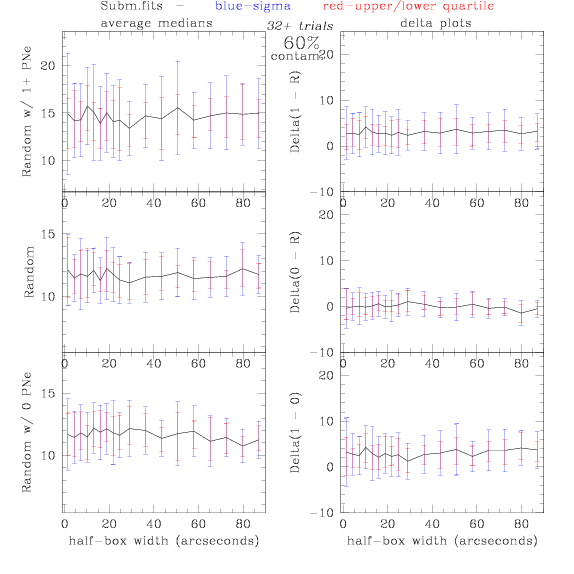

<-- less
contamination more contamination -->
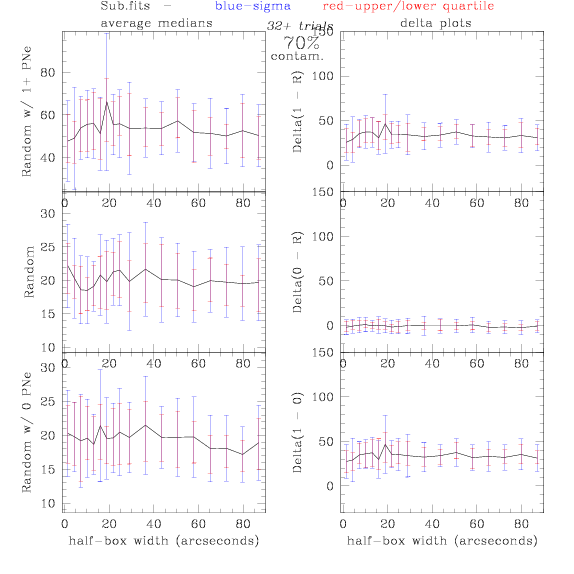
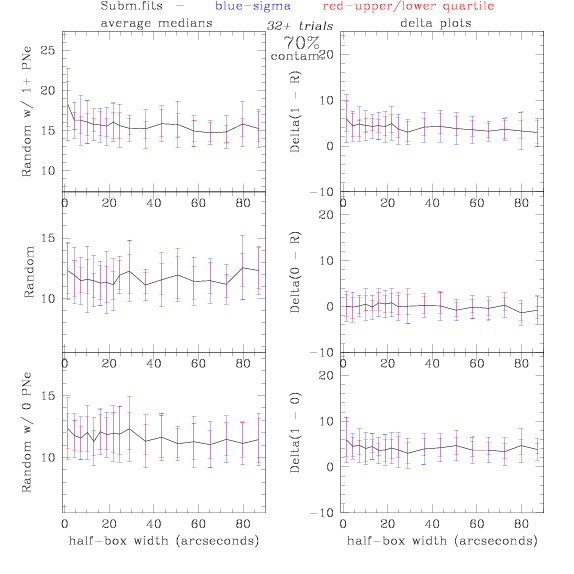

<-- less
contamination more contamination -->
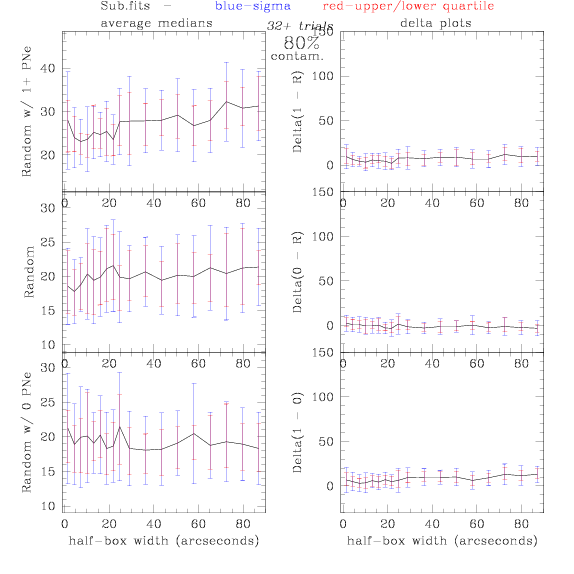
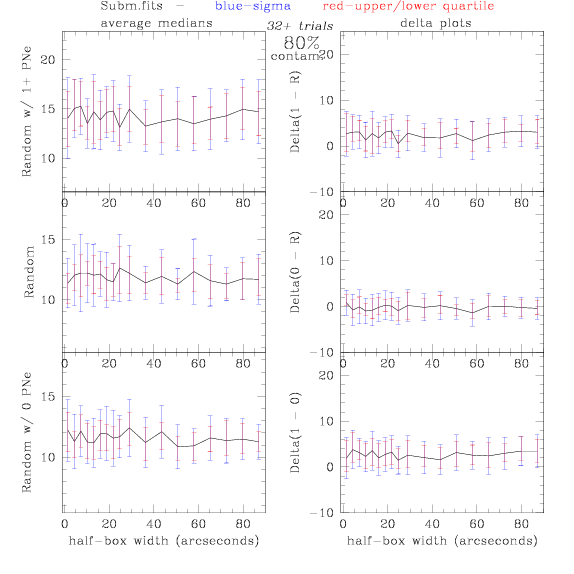

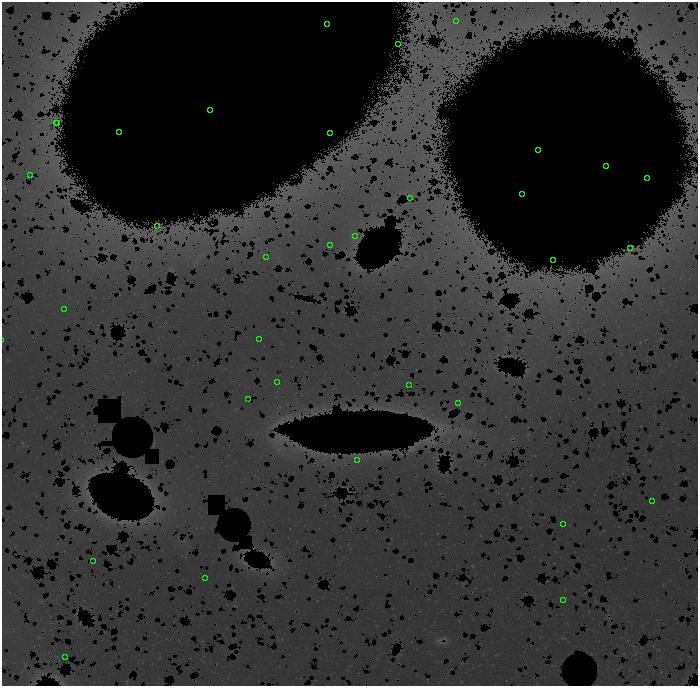
<-- less
contamination more contamination -->
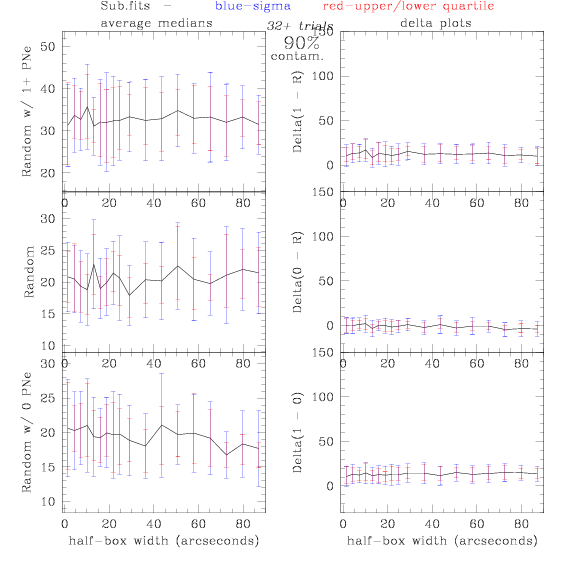
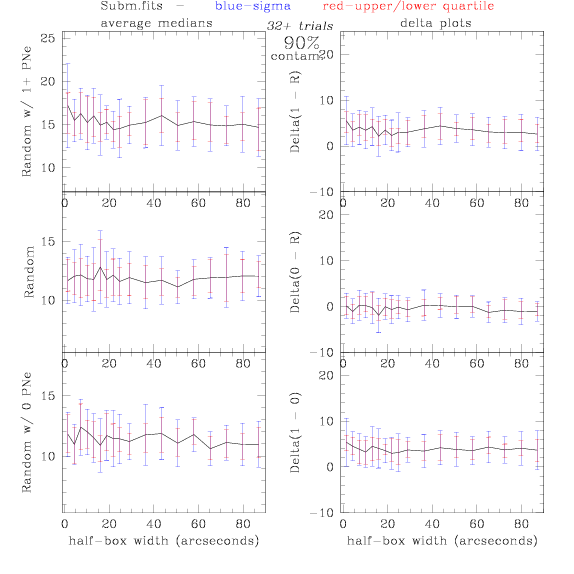

<-- less
contamination more contamination -->
back to 0% contamination