PNe
statistics, part 3
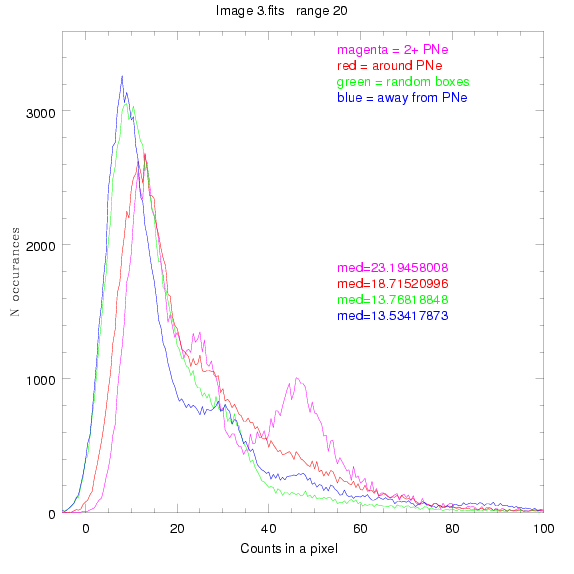
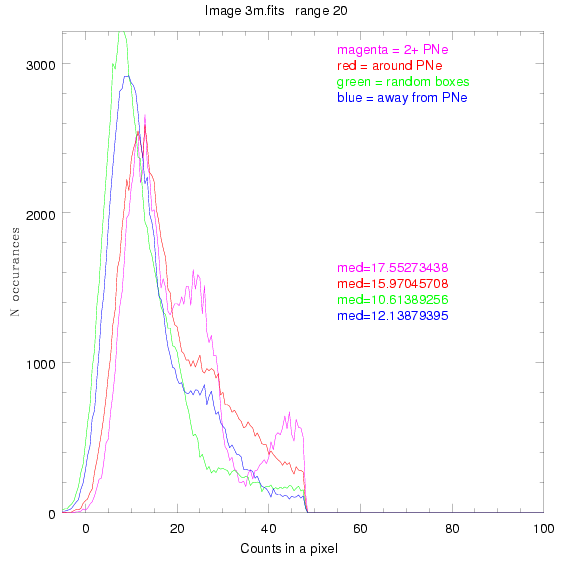
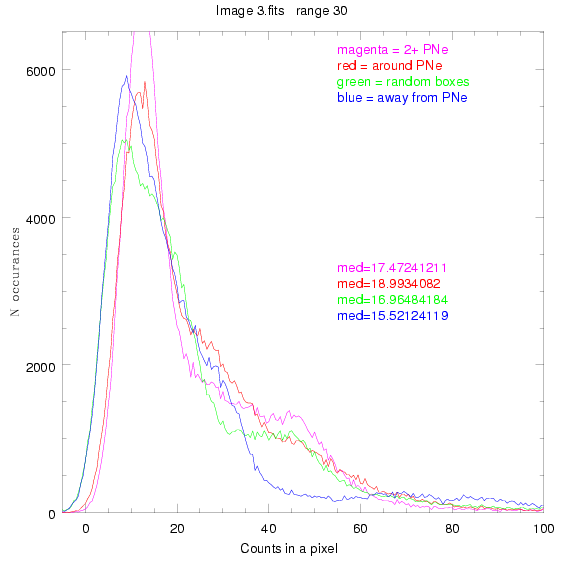
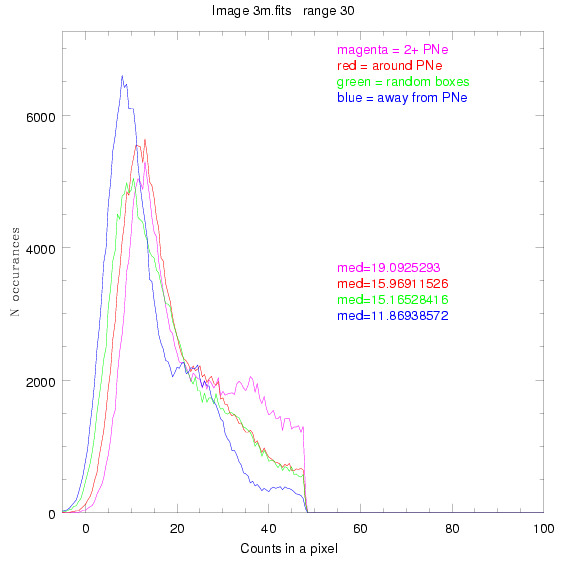
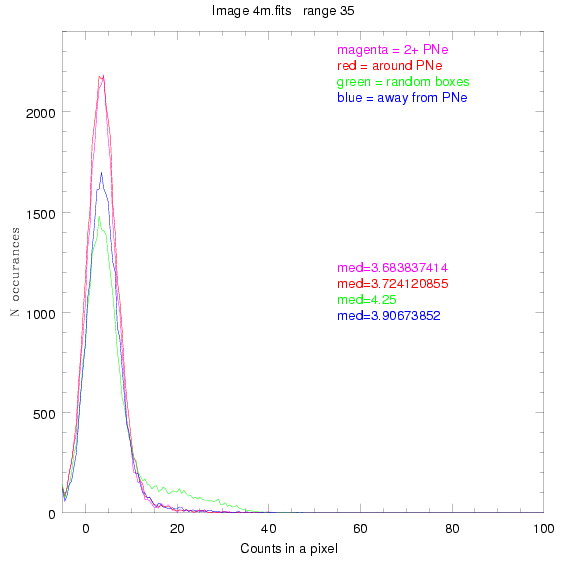
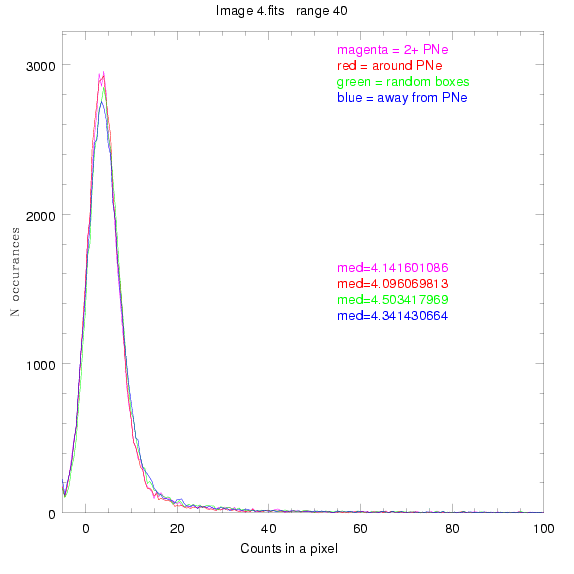
We are still attempting the same general idea as before, but now
looking at light distributions around PNe vs. not around them. As
before, we are breaking the image up into small square regions and
addressing the data in each of them.
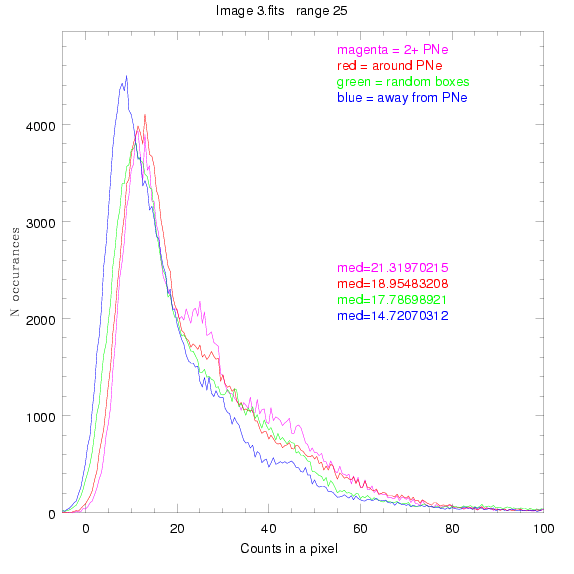
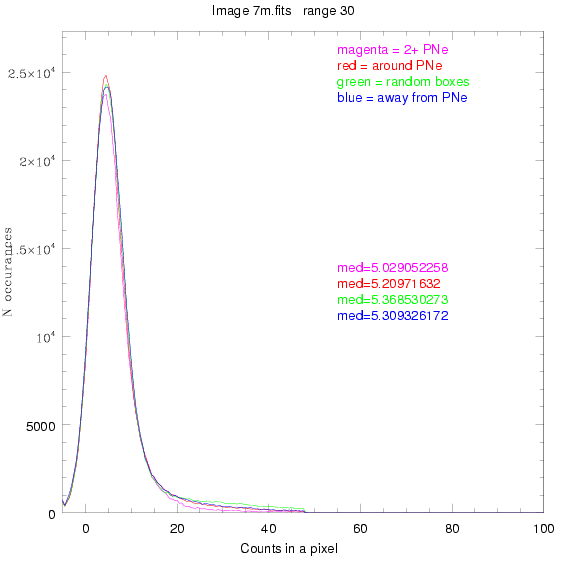
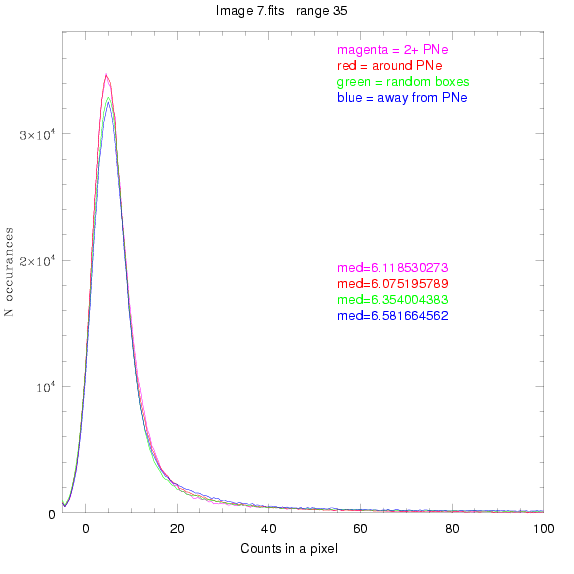
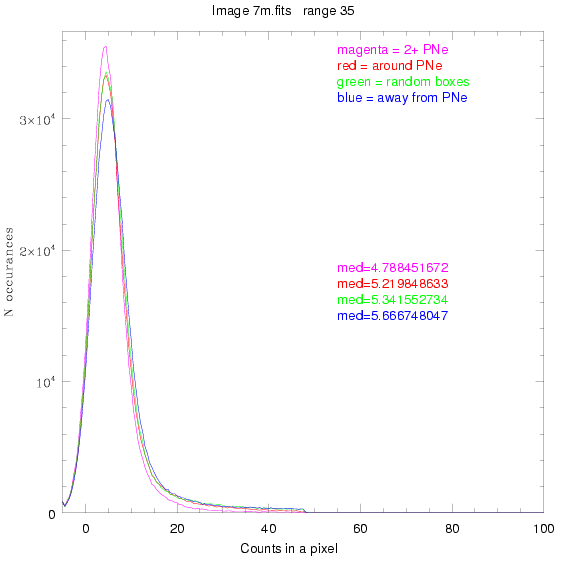
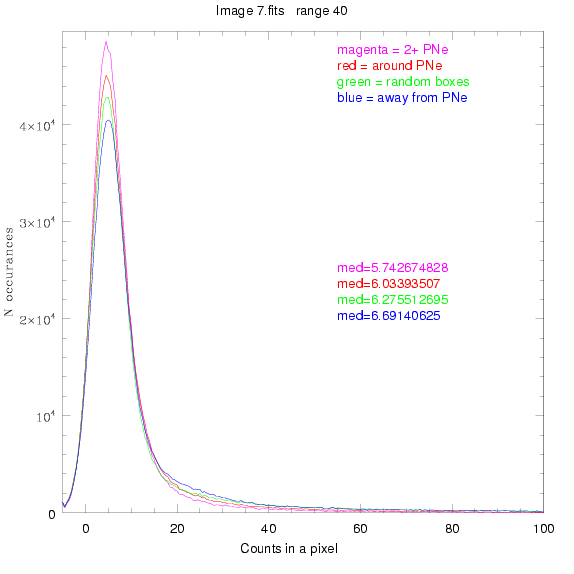
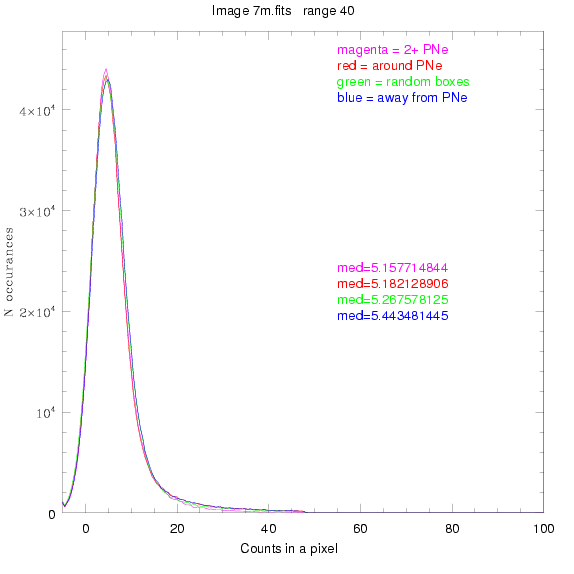
To select our regions we used three methods:
1) (magenta)
pick each region so that there are 2 or more PNe in it
2) (red)
center each region on a PN, so that there are as many regions as PNe
3) (green)
center each region randomly in the image, but only select as many
regions as there are PNe, for consistency
4) (blue)
select each region randomly but make sure that it contains exactly ZERO
(0) PNe
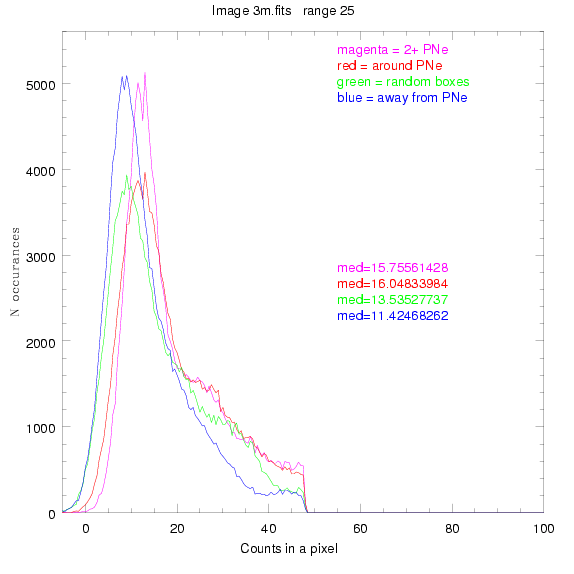
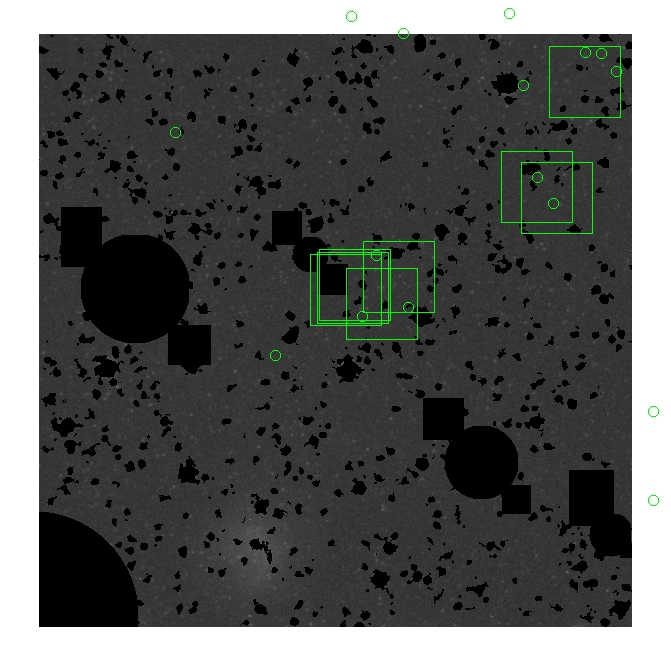
These three methods are applied on the masked and unmasked
images (side by side) with ranges (related to size of region) of 20,
25, 30, 35, 40.
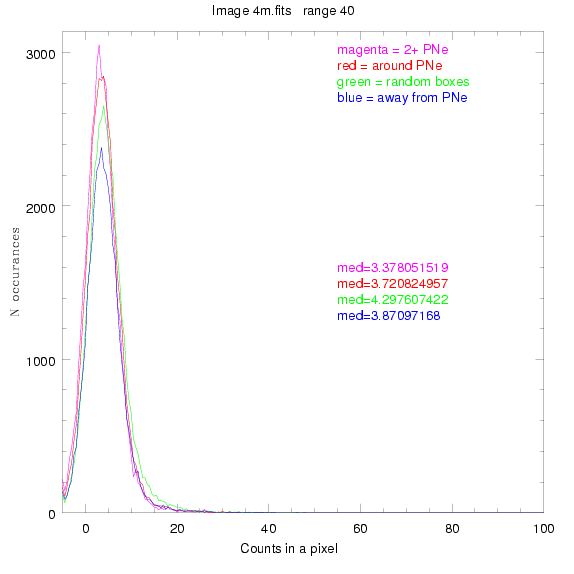
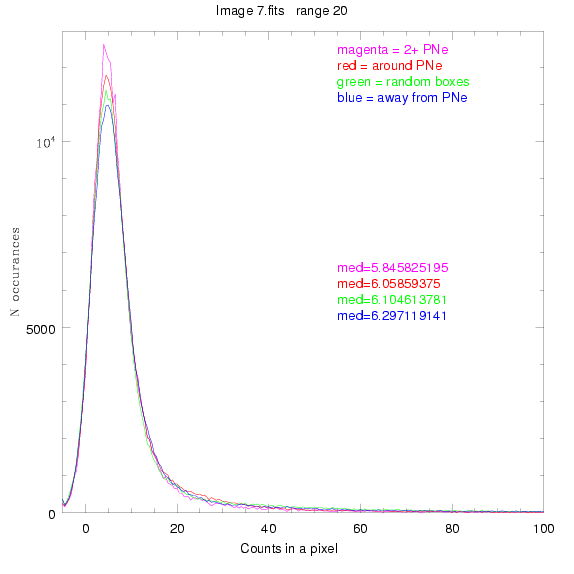
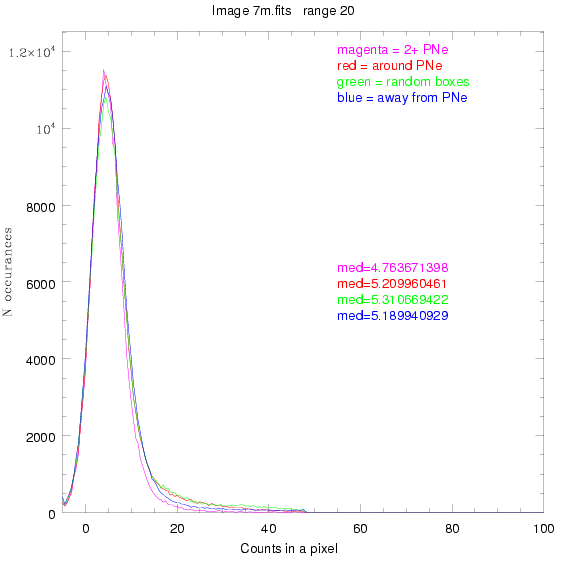
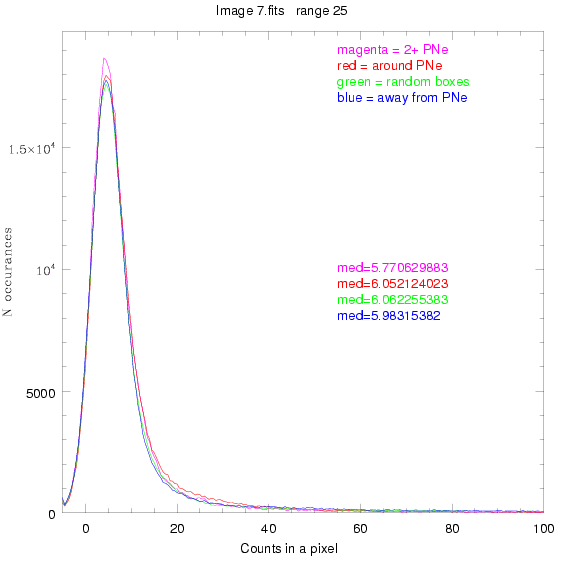
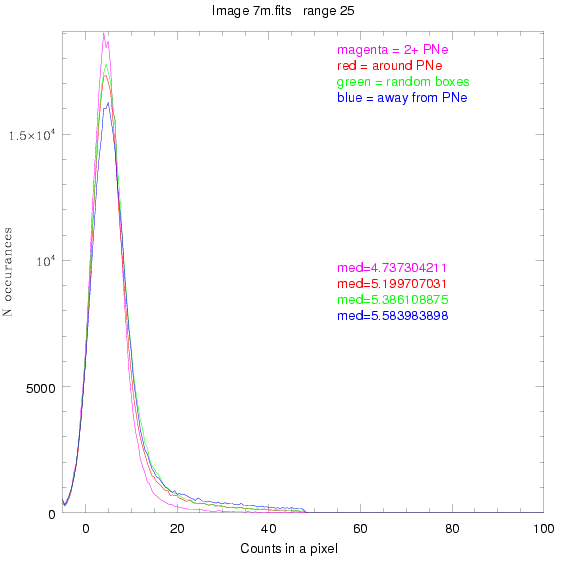
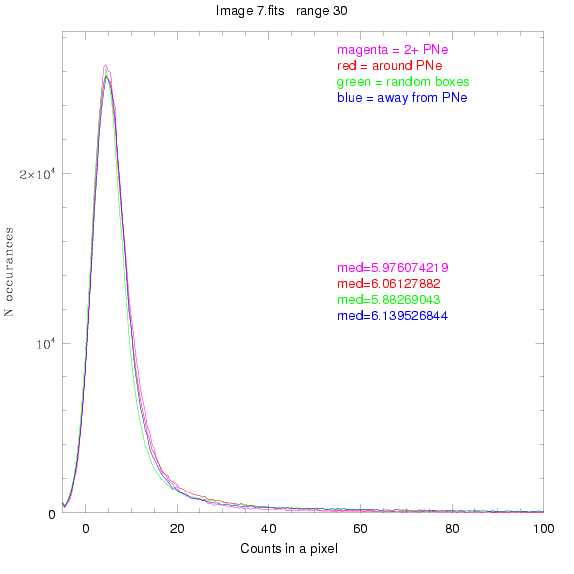
For each field (masked or unmasked), the median of each method was also
determined and is shown on the graph as "med=##"
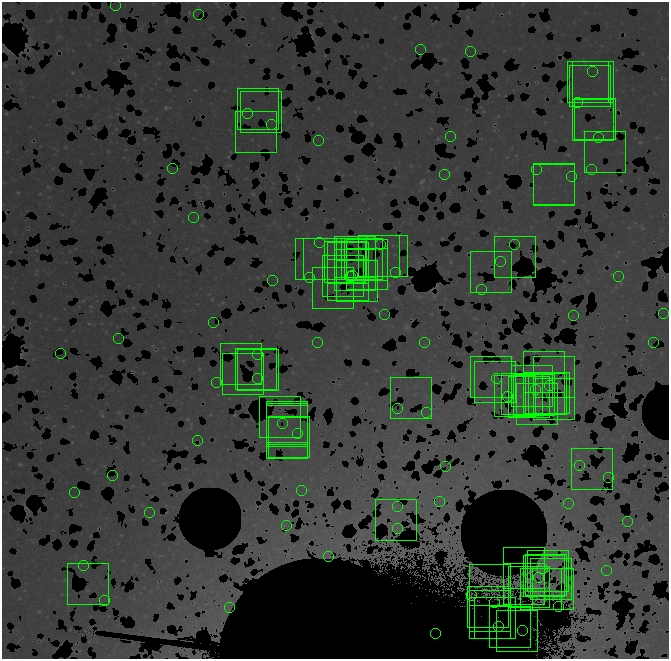
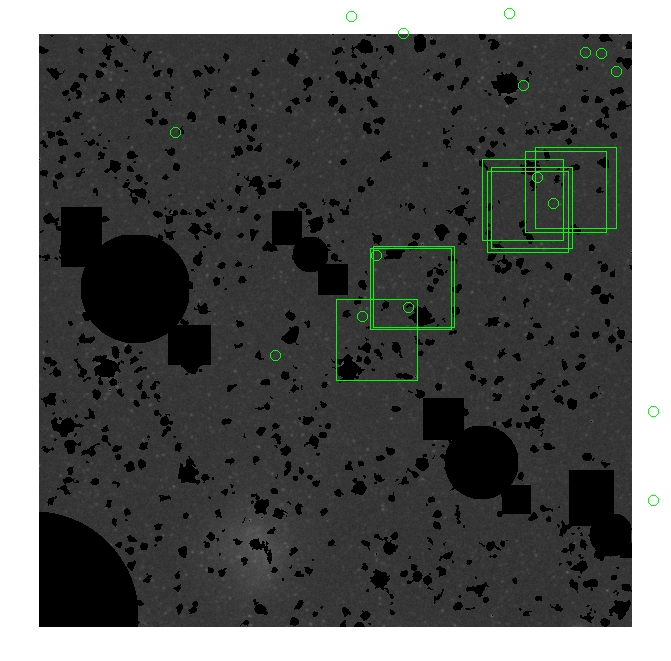
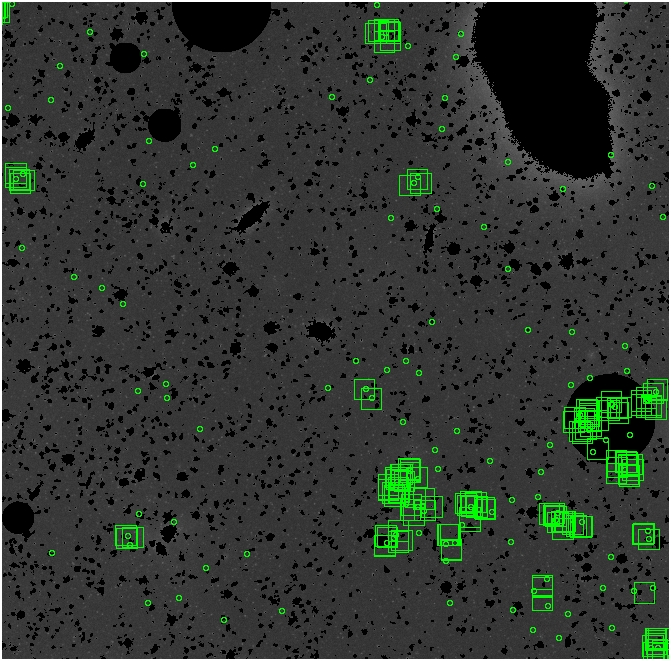
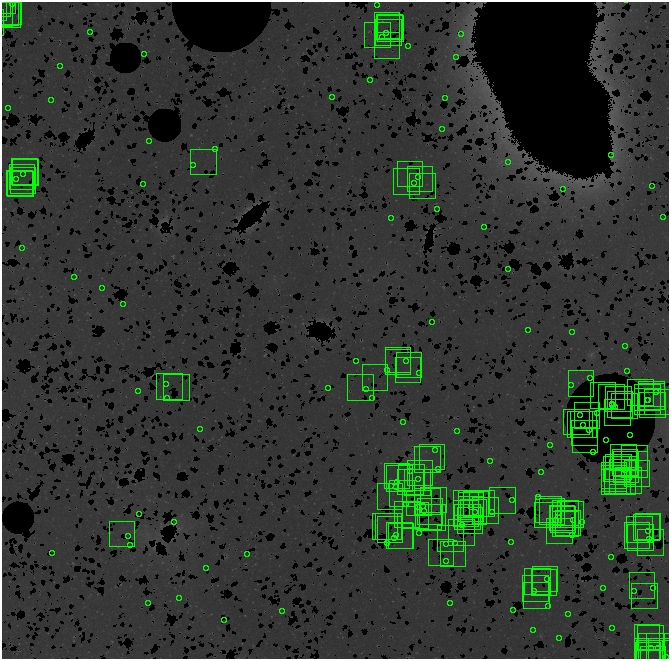
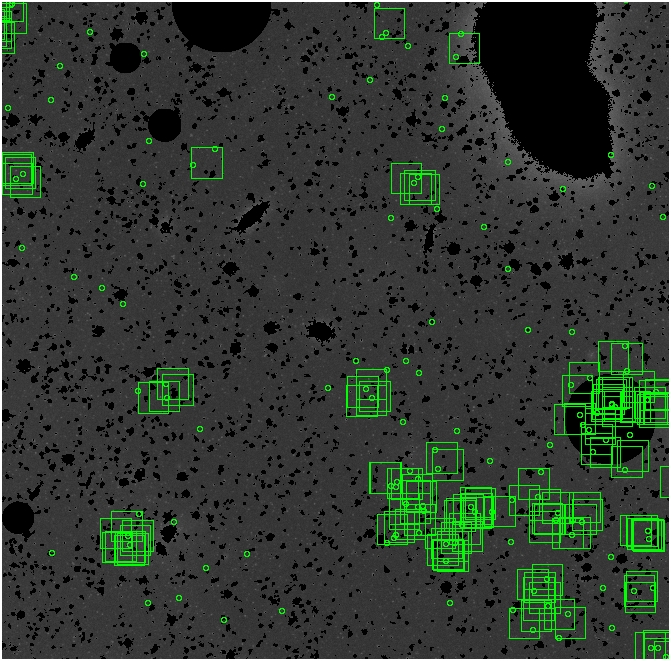
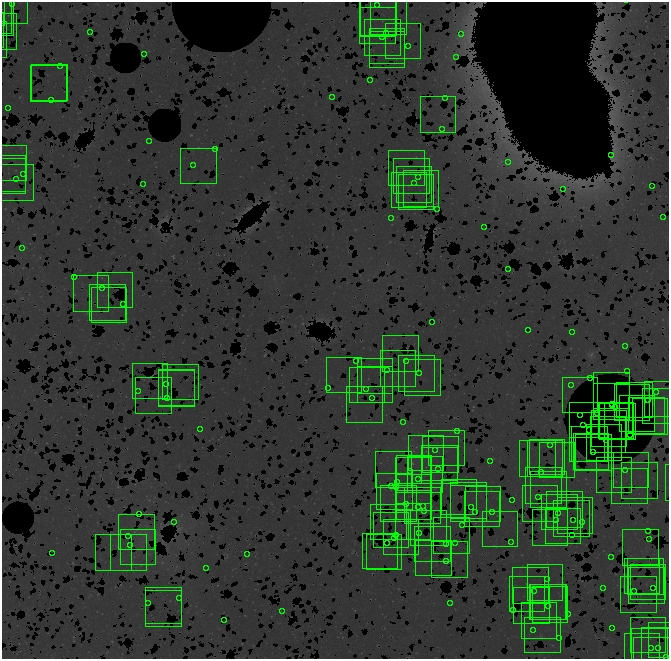
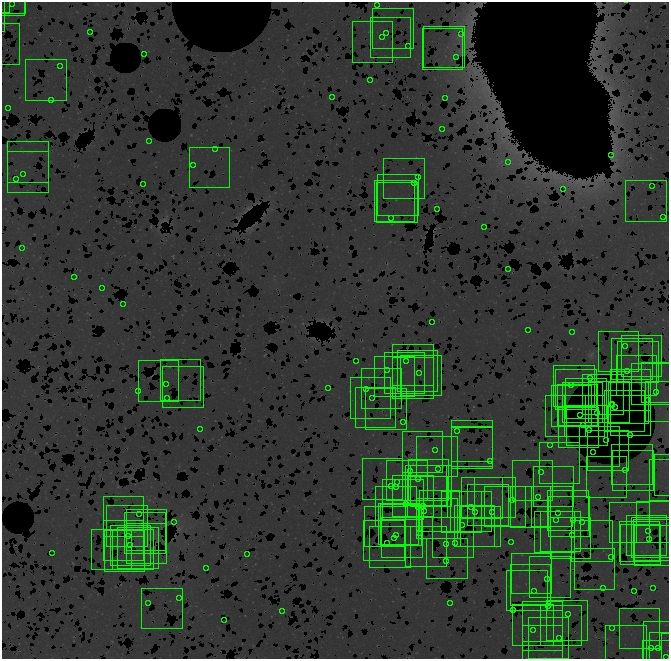
The masked images are masked with our best mask, down to u = 25, as shown on this earlier page.
These medians are collected and plotted as a function of range, shown at the bottom
of this page.
This was performed on 3.fits, 4.fits,
and 7.fits, using PNe catalog data from Feldmeier.
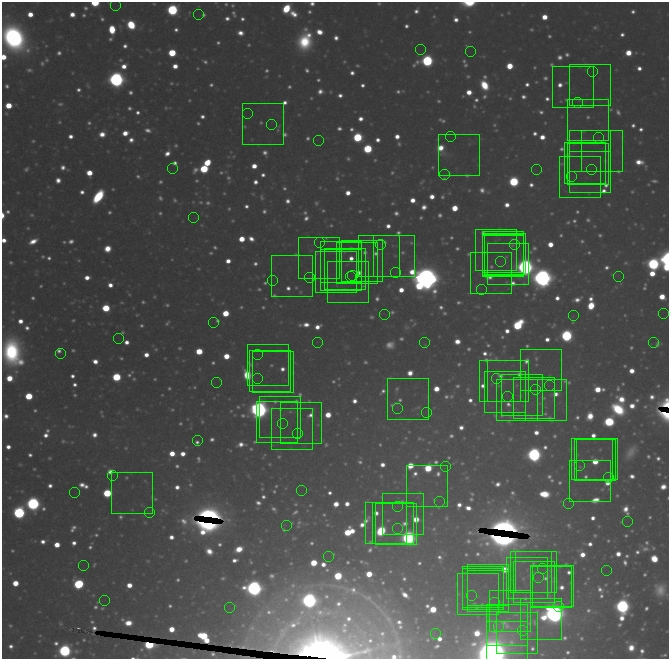
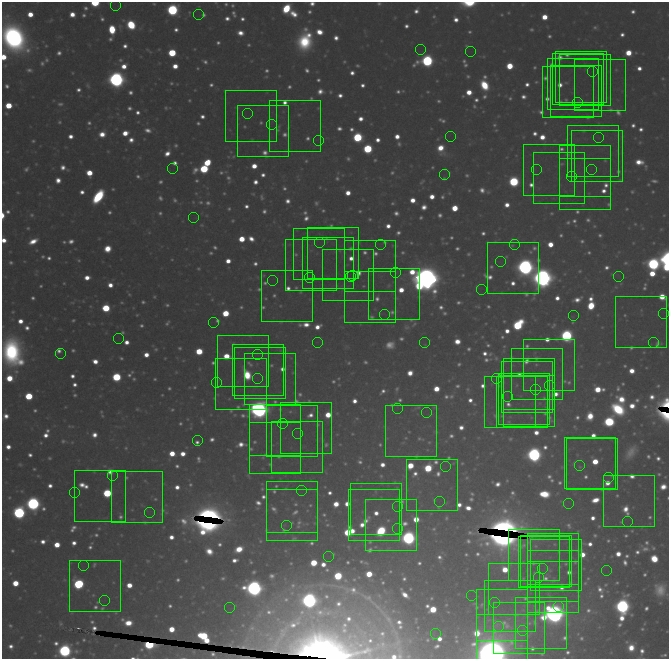
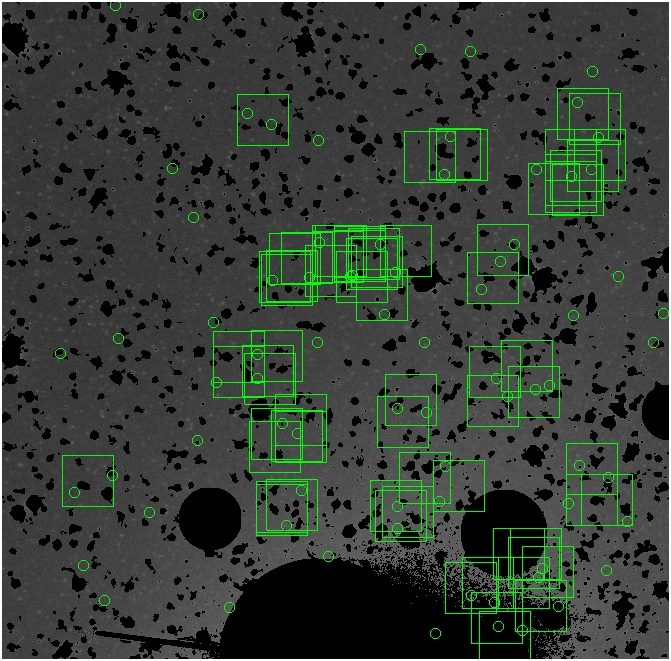
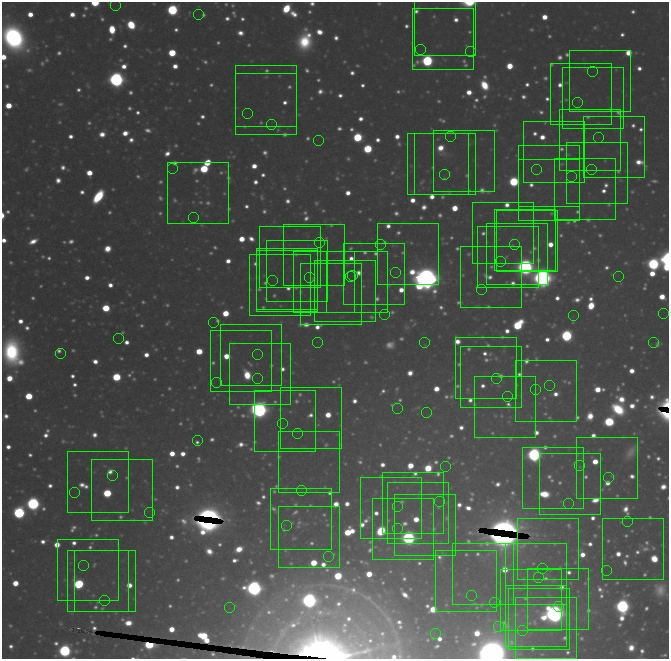
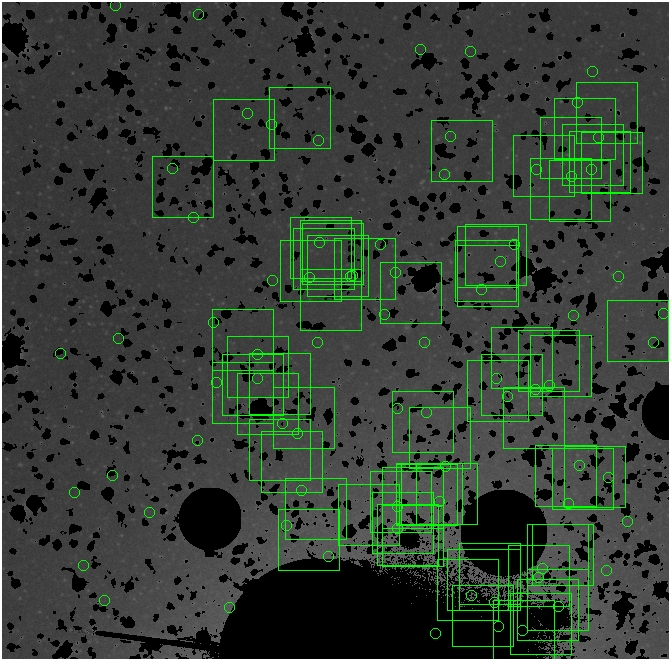
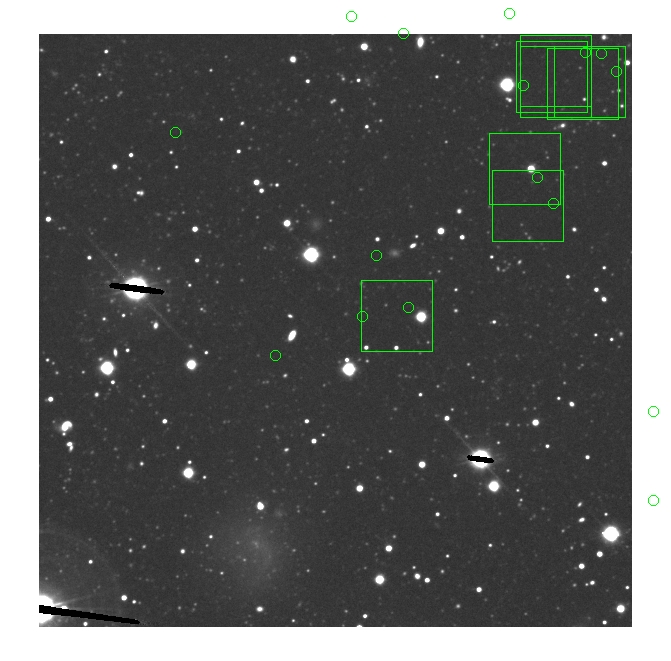
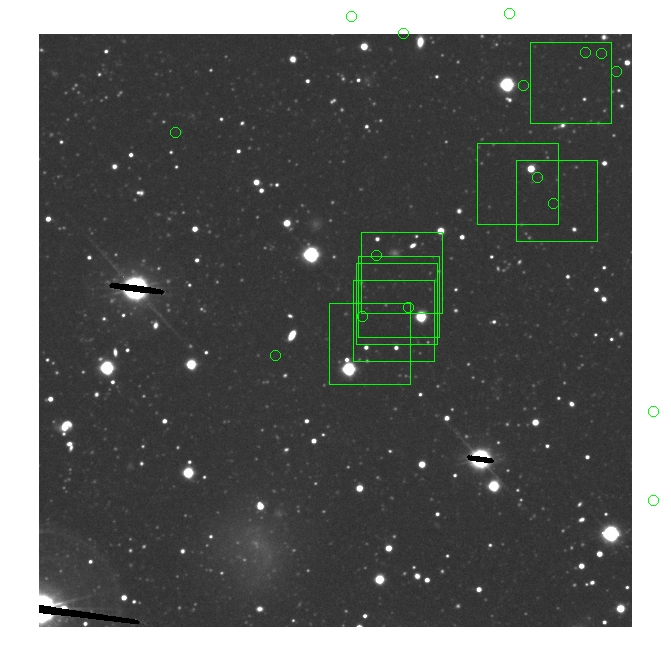
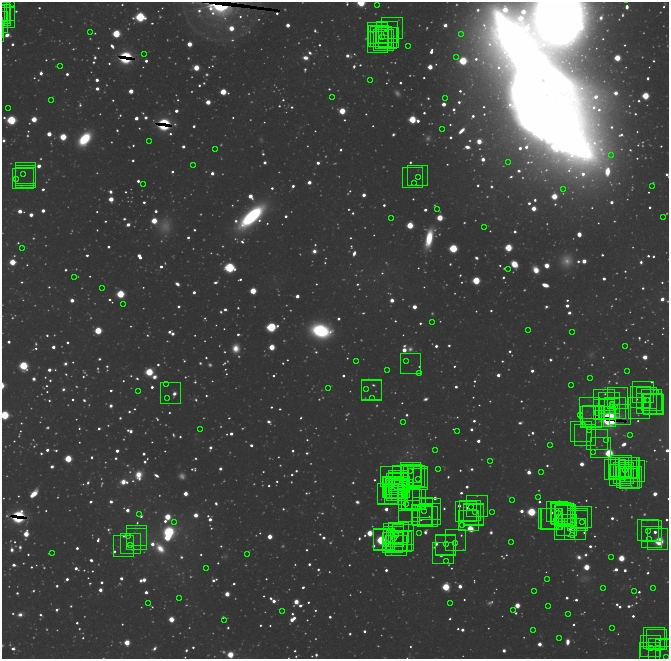
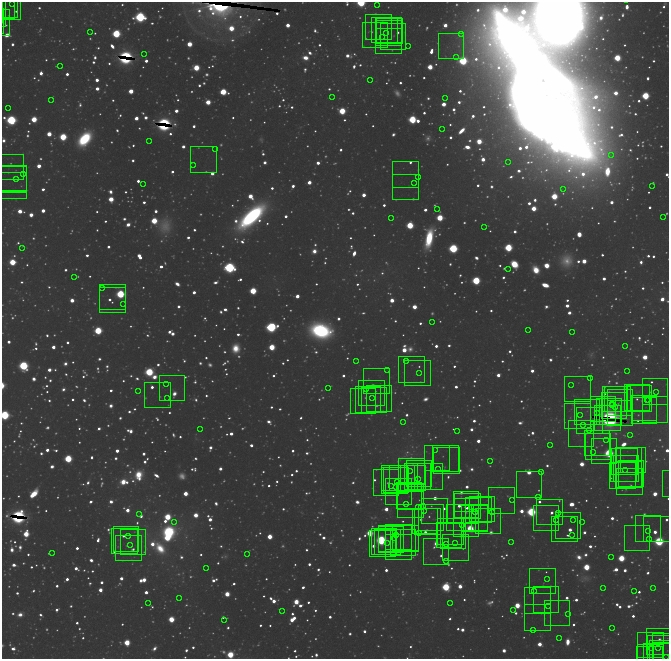
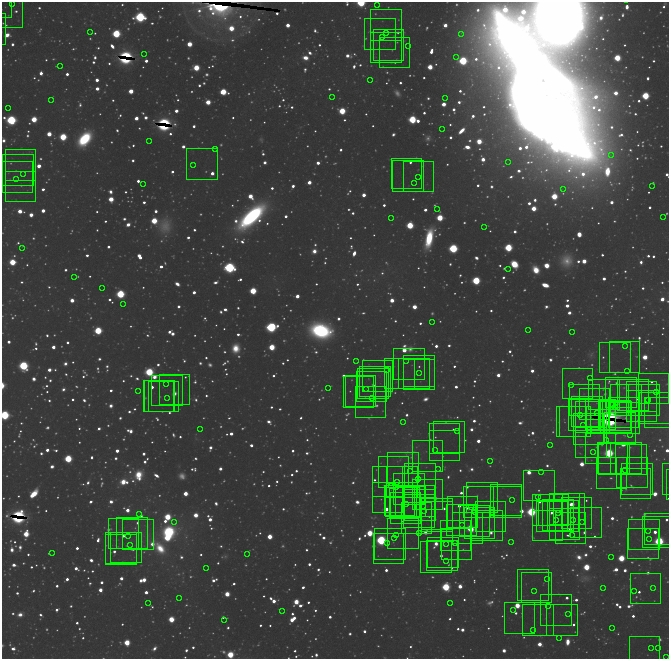
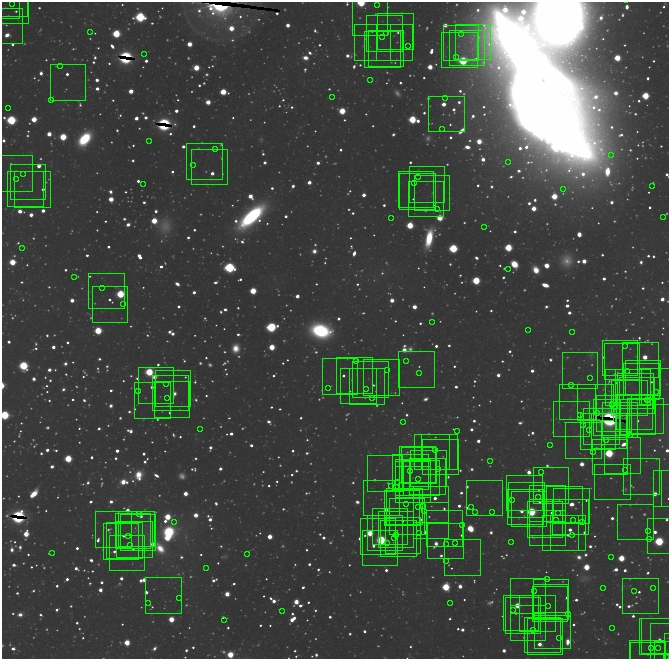
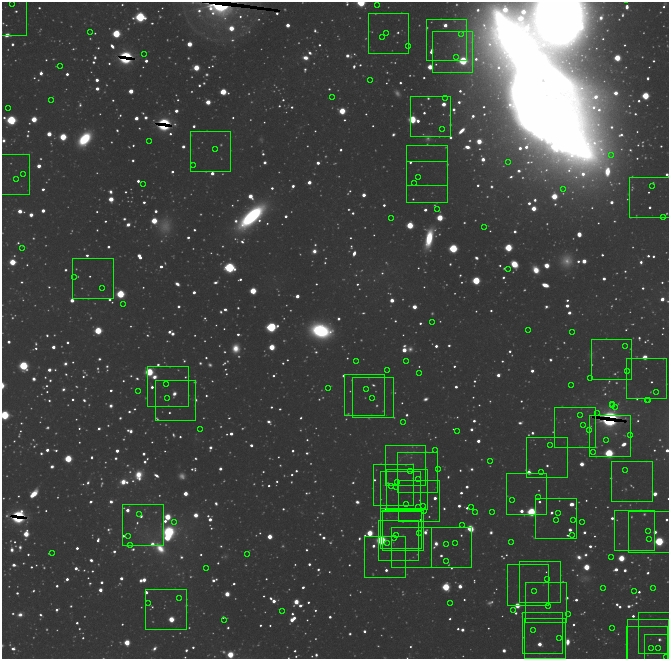
BIG DIFFERENCE: now, with the PNe >= 2 histogram, the curious humps
in the graphs are made clearer with images that are also included
showing where each histogram randomly chose its PNe >= 2 regions.
These follow each pair of histograms
code: PNe2.sm PNe2 <-- goes through and generates all the .ps files
as shown converted to .gif below and stores median values in meds.dat
PNe2.sm PNe2m
<-- takes data file meds.dat and generates bottom two figures
3.fits
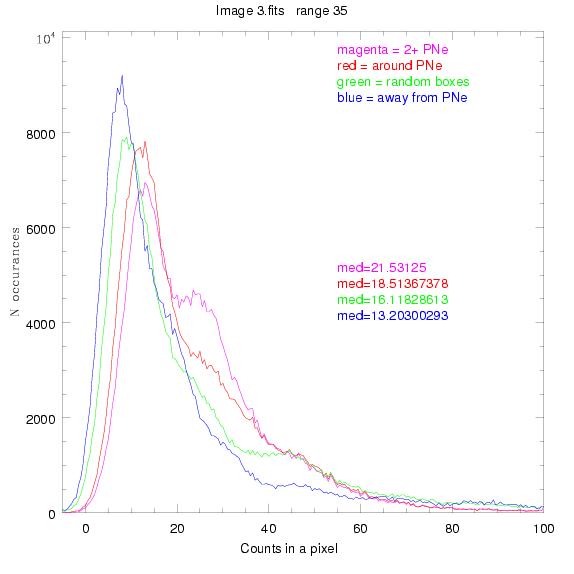
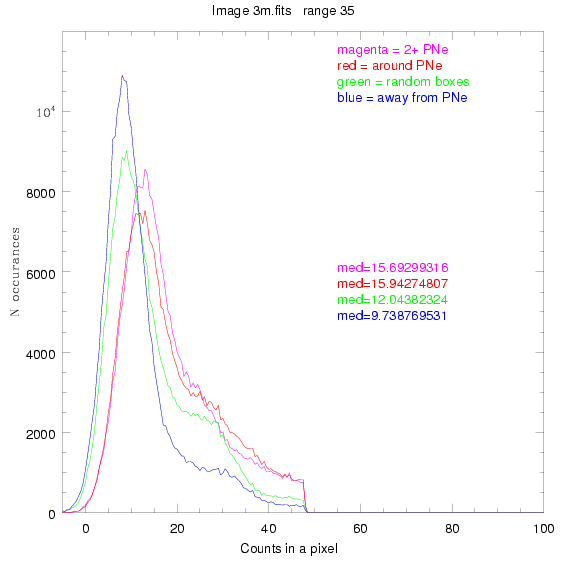
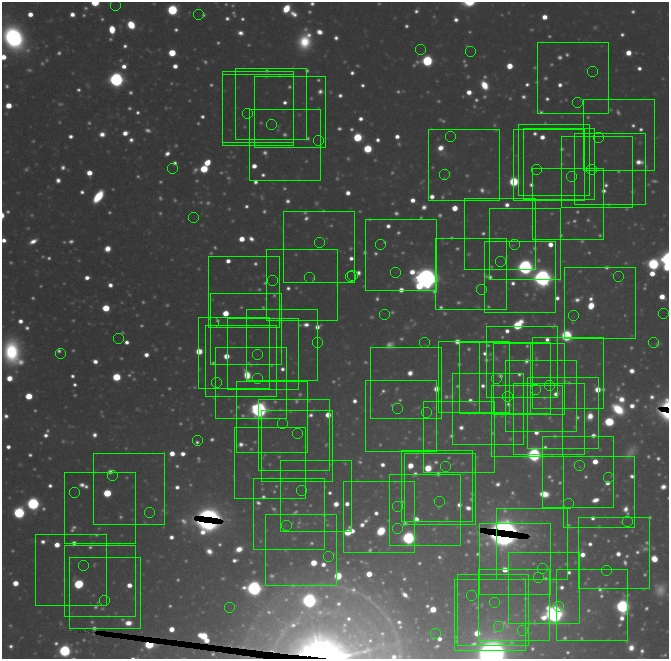
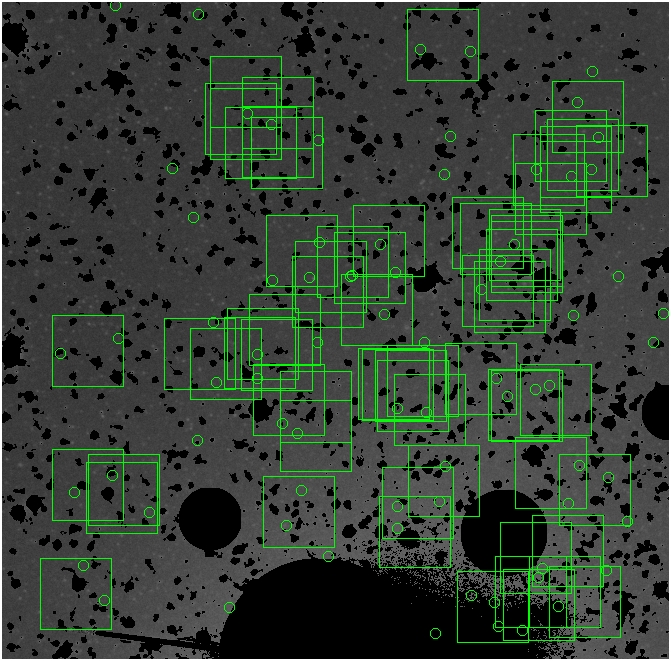
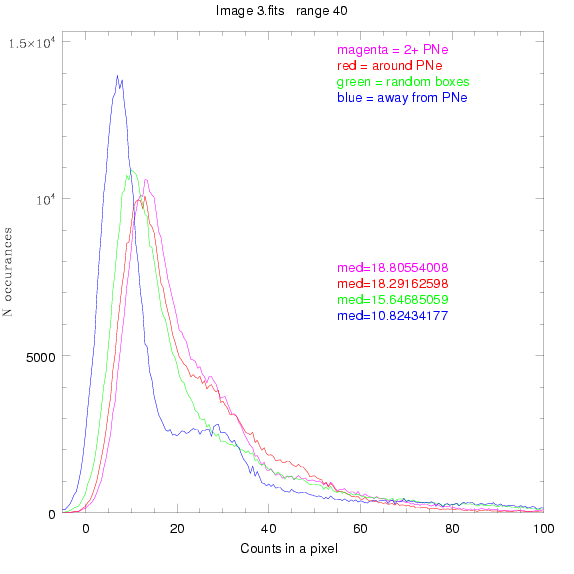
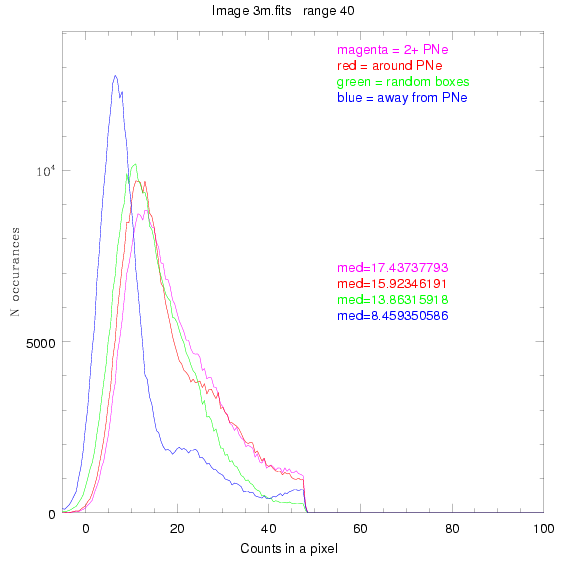
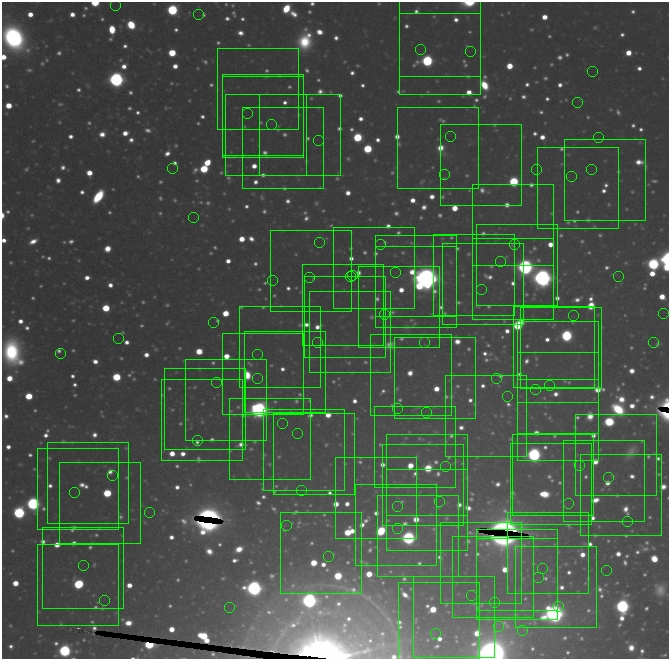

4.fits
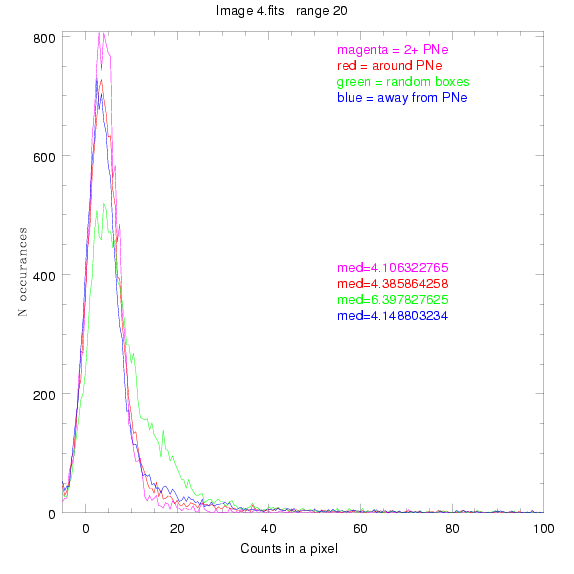
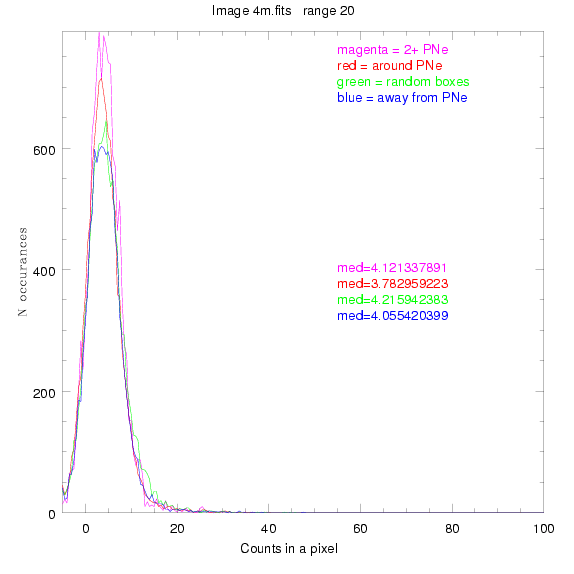
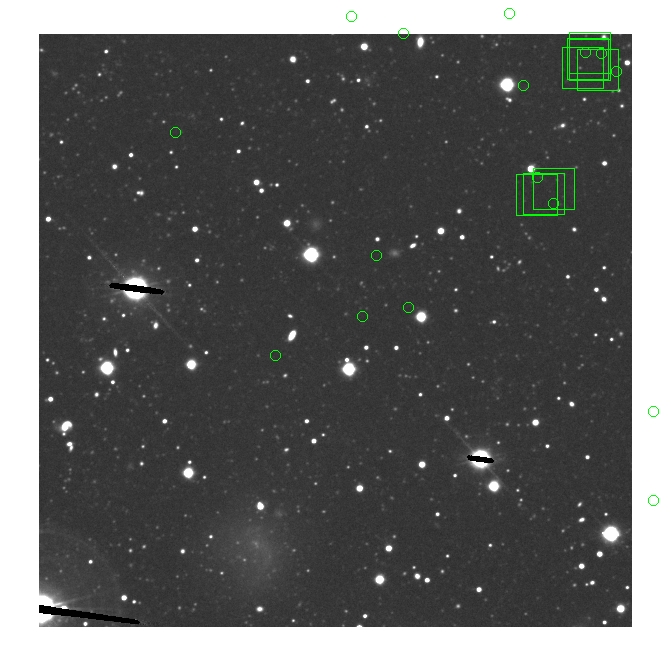
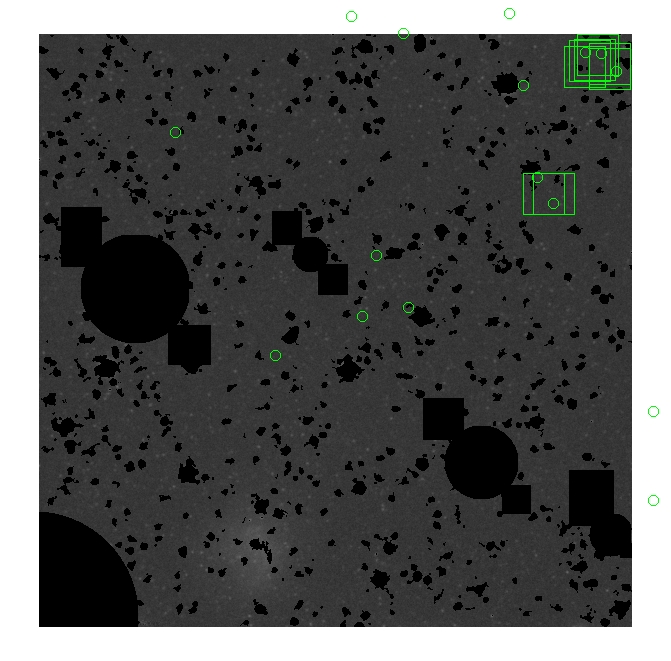
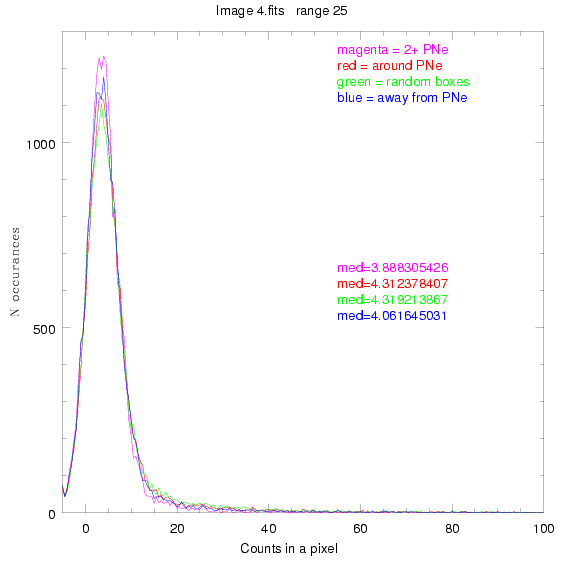
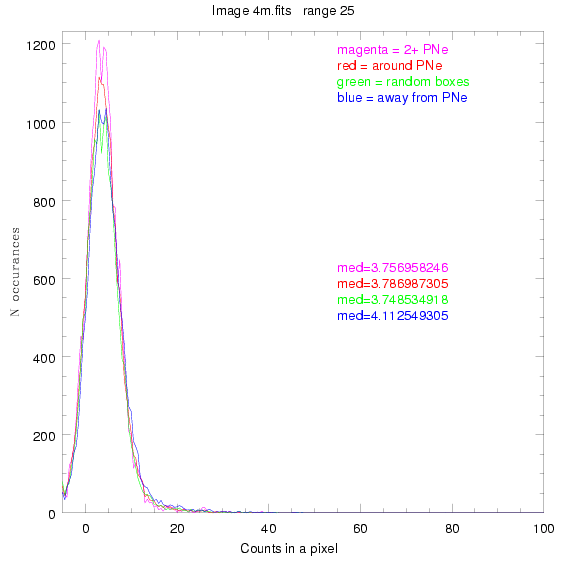
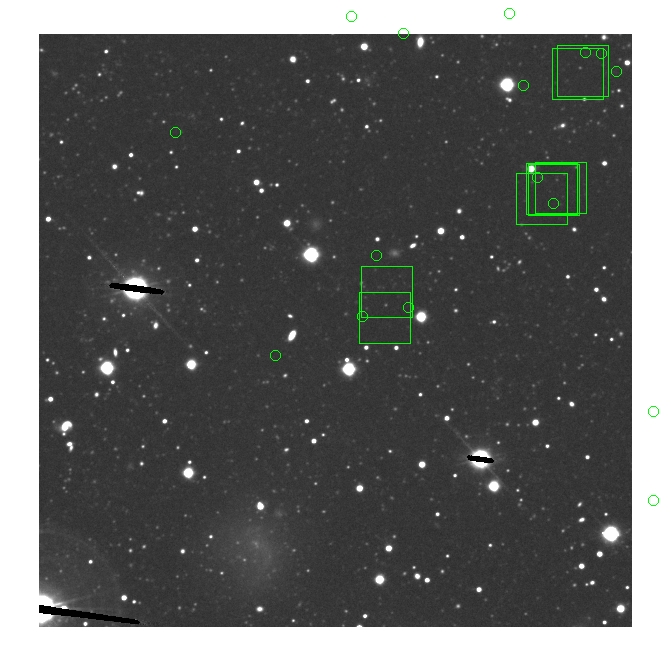
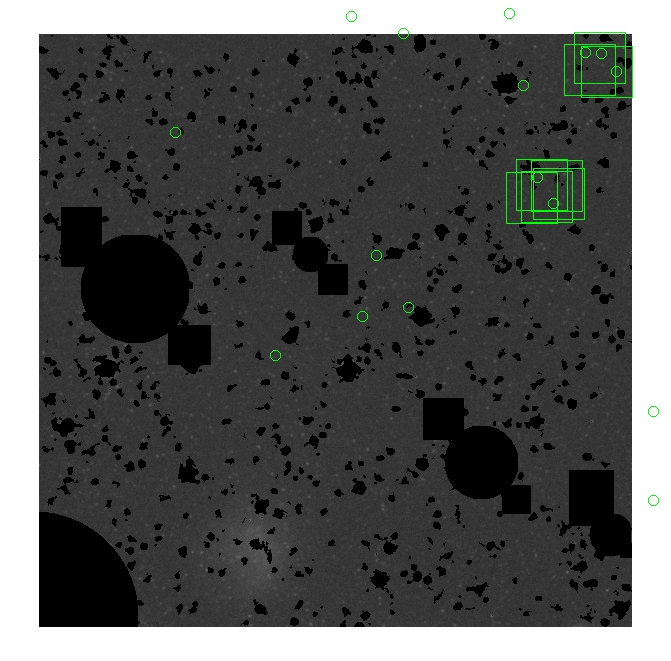
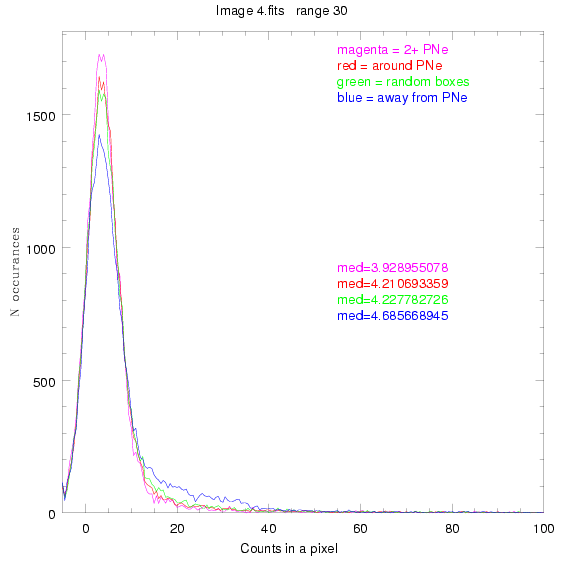
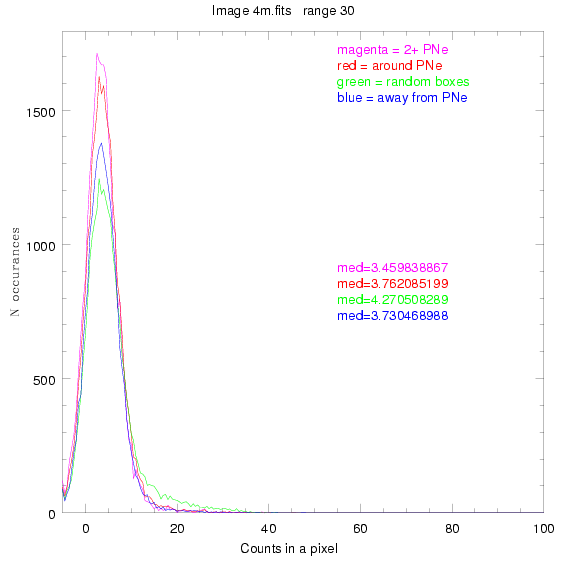
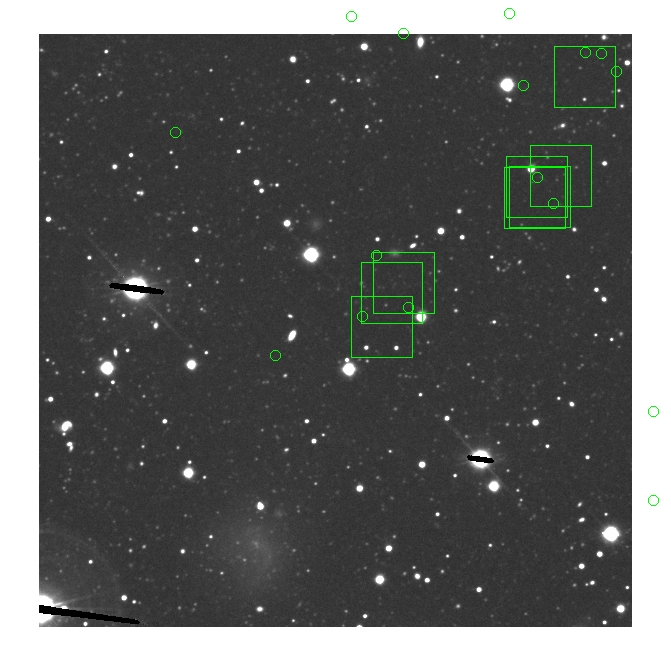
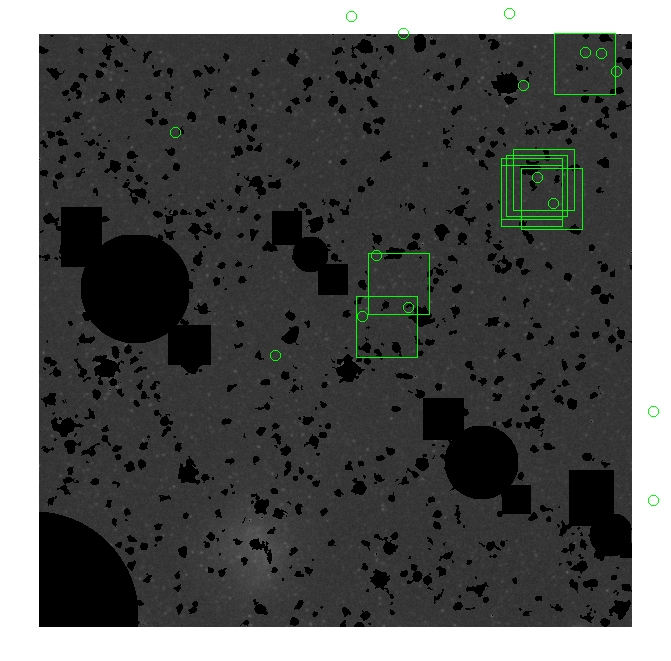
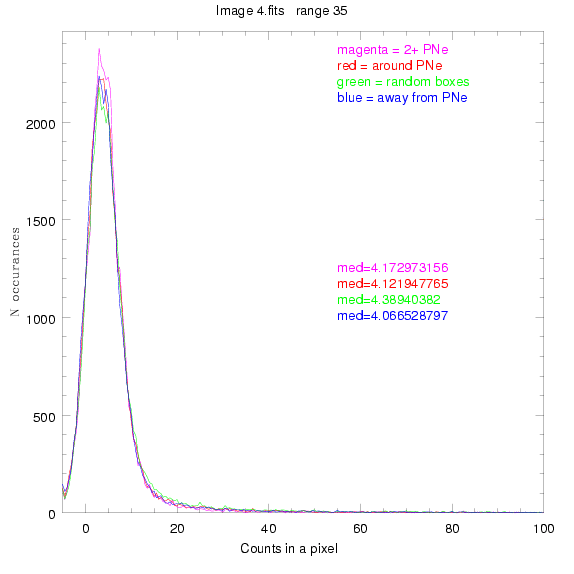
7.fits
all 574px wide
But there's more! what if we make sure the
boxes that have 2+ PNe in them are AT LEAST half a box width apart, to
prevent over-sampling?
Back to previous PNe stats page
Back to MCu stats page
Back to main Virgo page





























































