Final statistical analysis on PNe data
After much deliberation, experimentation, and trial and error, this is
the final procedure decided on to analysis/examine PNe data.
These methods are detailed below in what feels like too much detail but
should be helpful later on down the road.
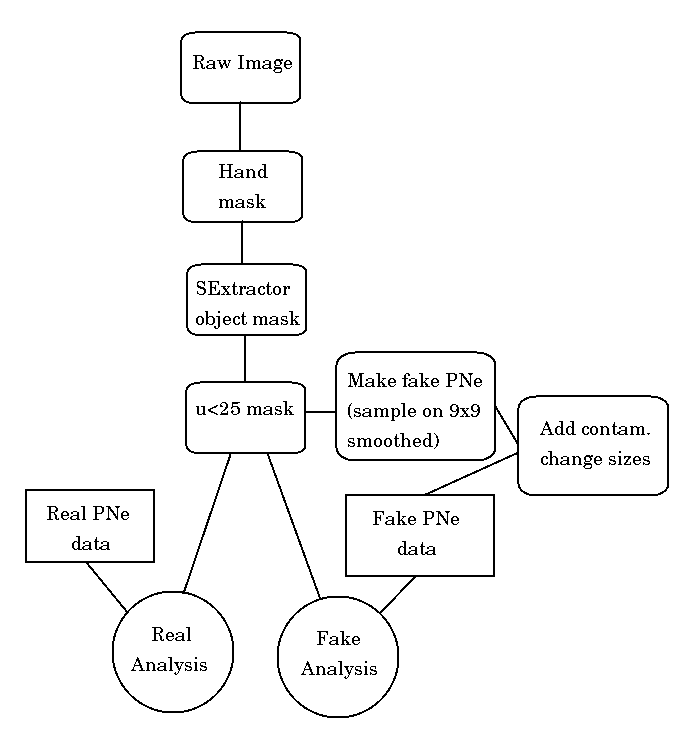
Here is an updated (9/21/07) flowchart for masking/etc:
Here's a menu/index of all the information contained
on this page (it is a lot):
Real Data: [PNe28 - click
for standard small individual summaries]
Details of analysis/code/methods
Results/graphs
Theoretical Data: [PNe26 -
click for standard small individual summaries]
Details of
analysis/generation/code/methods
Results/graphs
Comparing Real and Theoretical:
Comparisons/Over-plots
Re-Graphing (separate page) display
any/all
of the data for yourself
Analysis on Real Data:
Download the analysis code here: PNe28.sm
[renamed to PNe28.sm and main macro is PNe28, just for
consistency/clarity]
Download the graphing code here: PNe28Error.sm
[renamed also with the 28 for clarity]
If you want to actually DO graphing again, go to this re-graphing page.
The real data comes in 7 fields:
Aguerri's Fields (Core, Sub, FCJ, LPC) [PNeA.dat - Field, PN#, RAh,
RAm, RAs, DECd, DECm, DECs, M5007, NOTES]
Feldmeier's Fields (3, 4, 7) [PNeF.dat - Field, PN#, RAh,
RAm, RAs, DECd, DECm, DECs, M5007, NOTES]
If you need these files, they can be found in my
home directory, ~/Virgo/FeldPNe/ under the above file names.
As the analysis is done on each field, it only opens the appropriate
data file, as there is overlap between some fields, but we do NOT
want overlap in the data files.
All box selections are written out to data files as part of this
program:
Boxes with 1+ PNe: 28.1.Im3.fits.R1.wfound.dat [RA
DEC RANG]
Boxes with 0 PNe: 28.1.Im3.fits.R1.0found.dat [RA
DEC RANG]
Boxes selected randomly: 28.1.Im3.fits.R1.Rfound.dat [RA
DEC RANG]
1 is trial number that goes from
1-20
3.fits is the field
(unmasked field 3)
R1 is statistics box range
(half-width) of 1 (3x3 px)
Trials are set by a small file called i.i, which contains just a
single number on the first line. This file must exist for PNe.sm to run. The
number it contains is the trial number PNe.sm will perform when it is
run. That is, if i.i is
1, PNe.sm will run trial
#1 when executed,
and will, as soon as it has started, change i.i to 2, so that the next
instance of PNe.sm
(presumably you're running a whole bunch on
the cluster) will start on the next trial number.
Other important small files are: v.v,
PNe.param.
v.v -
this file sets the version number, and should usually match the folder
it's in - PNe28 should have v.v
set at 28.
PNe.param - this file tells the
code whether or not to allow boxes to overlap each other. In version 28
(this version), this file is no
longer needed as boxes are always
allowed to overlap, but it still is important in other versions.
Selection of fields to use and ranges for statistics box size are set
in the actual code (admittedly sloppy, but effective). If no changes
are made to the lines defining them, all fields will
be used over all ranges. If you want to select certain ranges to run,
simply add
a line like: set raO = { 1 3 5 7 9 11 13 15 }
and only the ranges stored in that vector will be used. Similarly, the
images to be
run are stored in a vector, except they are slightly
more complicated, as there is the issue of side-by-side images now so
that we
can read from the "MM" masked image. To run all
images, the code looks like this: set imO = { 0 1 2 3 4 5 6 7 8
9 10 11 12 13 }
and if you want to only run certain ones, you must
compare that vector of numbers to the list of images directly below it
in the code.
Here is a handy small table to show how the numbers
correspond to the fields:
0
|
1
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
9
|
10
|
11
|
12
|
13
|
| 3.fits |
3m.fits |
4.fits |
4m.fits |
7.fits |
7m.fits |
Core.fits |
Corem.fits |
Sub.fits |
Subm.fits |
FCJ.fits |
FCJm.fits |
LPC.fits |
LPCm.fits |
So use the set imO = command to
choose whichever numbers for the fields you wish to run.
pickgoodbox is a macro in PNe.sm that selects a random
box (center) with constraints:
box (center) must be within the limits of this
image, less the size of the statistics box (so that a statistics box
can collect pixels all
around this center)
box (center) must be on an unmasked pixel when
looking in the "MM" masked image [i.e. 3mm.fits] - the image with
bright star
mask and SExtractor/MonteCarlo
mask, but NOT surface brightness cutoff (used for theoretical data
generation)
After a box has been selected, it is tested to see if there are any PNe
in it, by calculating the distance between all PNe in the catalog
and the box center. If any PNe are found, the box's
coordinates are added to the vector that will eventually be written out
to the
*.wfound.dat
file and the pixels inside the box are added to the vector of pixels
from boxes with PNe in them.
If the box contains 0 PNe, it is added to the storage of boxes without
PN, the coordinates to the *.0found.dat
and the pixels to the
appropriate vector for storage.
Random boxes are generated by simply querying the pickgoodbox macro and
unconditionally using the output as the box center.
The number of each type of box is set to be the same as the number of
PNe observed in the field, and these numbers are preset
and hard-coded into the code.
After boxes have been selected, coordinates saved/written out, and
pixel values concatenated, we move into the histogramming and
graphing section.
The stored pixel vectors are histogrammed and the graphs are outputted
as 28.1.Im3.fits.R1.ps.
One of these graphs contains all three
types of boxes, each in a different colour, with
their medians also displayed on the graph.
The computed medians are stored in a data file called which is drawn on
later by PNeError.sm to
graph up the error bars and
range-dependent plots. This file is 28.1.Im3.fits.R1.med [image
range 1+PNeMedian 0PNeMedian RandomMedian].
PNeError.sm is the program
that will combine all of these *.med
files from all ranges for each field.
At the beginning of the code there are vectors to set of running
parameters. Similarly to PNe.sm,
they are:
For range: set ra = { 1 3 5 7 9 11 13 15 25
35 45 55 } (these should match the parameters used in PNe.sm or else it will try to
read files that do not exist)
For fields: set im = { Core.fits Corem.fits
FCJ.fits FCJm.fits } (again, you should be sure that the first
part of the analysis has
been run and *.med files exist
for the selected fields)
This code is robust enough to know to skip files when they do not
exist, but this can cause strange things to happen on the output graphs.
If the output graphs have points with value 0 or
huge error bars, this is probably happening. To make sure it's reading
all the files, watch
the terminal output as you run the program and be
sure all trials (1-20, or whatever you've done) are being read in.
Currently, this program is hard-coded to read in trials (28.trial_number.*med) numbered
1-20. To change, redefine the variable itt near the
beginning of the code. Whatever itt is
set at will be the maximum trial number, starting from 1.
After this program reads in all of the *.med files, it writes out *.stats files which follow the
naming convention: 28.0.Im3.fits.R1.stats
and has
the following format: (the top row is in the actual
data file, as a comment (#) to help remember what the entries mean)
#1+PNe (wPNe)
|
#0PNe (woPNe)
|
#Random (rand)
|
#Delta(1-R) (Dw)
|
#Delta(0-R) (Dwo)
|
#Delta(1-0) (Dwwo)
|
Mean
|
Mean
|
Mean
|
Mean
|
Mean
|
Mean
|
Sigma
|
Sigma
|
Sigma |
Sigma |
Sigma |
Sigma |
Kurtosis
|
Kurtosis |
Kurtosis |
Kurtosis |
Kurtosis |
Kurtosis |
Median
|
Median |
Median |
Median |
Median |
Median |
Lower
Quartile
|
Lower
Quartile |
Lower
Quartile |
Lower
Quartile |
Lower
Quartile |
Lower
Quartile |
Upper
Quartile
|
Upper
Quartile |
Upper
Quartile |
Upper
Quartile |
Upper
Quartile |
Upper
Quartile |
Finally, the program generates those plots that we've spent so much
time staring at, with the 6 sub-plots, error bars, Medians and Deltas as
functions of range, that so nicely sum everything
up. These plots are named 28.0.med3.fits.we.ps
and also converted to gif files as
28.0.med3.fits.we.ps.gif.
The values from the above table (data file) are used in plotting up the
data, somewhat obviously.
The PNeError.sm program ends by copying these new *.gif files to ~/Virgo/Web/PNe28/ folder and /astroweb_data/web/steven/PNe28/.
back to menu
Results of Real Data Analysis:
Click
here for the separate PNe28 page for
individual graphs
Click here to see the comparison graphs at the
end
between the Real method and the Theoretical method.
Do you like graphs? Want to make some more? Check out the Re-graphing page.
back to menu at top
Generation of and Analysis on
Theoretical Data:
Generation:
We now endeavour to simulate PN positional data to analyse identically
as above to determine what sort of results to expect. In
/packmule_raid/steven/PNe8alpha8/
there are programs which generate theoretical catalots of PN positional
data. These programs are
PNe8alpha8.sm
and subs.sm.
Both of those programs are available for downlad: PNe8alpha8.sm subs.sm
In PNe8alpha8.sm, there was a very
recent modification made to write out data 1 column at a time as the
image was scanned, in order to
distribute the job across the cluster and actually
finish it (without the memory getting very upset). Most of the fields
were not done this way,
but 7 and Sub were, as they were the largest.
subs.sm is rather old and clunky... I wrote it earlier in the summer
before I knew how to make variable names contain variables in them...
So,
rather than one set of commands to
subsample/contaminate, there are 7 nearly identical ones except with
slight changes for each field. I might
re-write it soon, if there is time this summer.
PNe8alpha8.sm is a program
that generates a massive catalog of PN positions (~1+ million for most
fields) based on their assumed correlation
with light. Using the "MM" mask, comprised of the
bright star hand mask and the SExtractor/Monte Carlo mask, PN positions
are correlated
with light.
Smoothing on the input image is set by the vector ranges
in the beginning of the code. Before it was realise how excruciatingly
long this code takes
to run, it was thought that it would be feasible to
run multiple smoothing sizes or images in the same realisation. It is
not, but the vector set up still
remains for many of the running parameters. Thus,
for 9x9 smoothing (as desired), simply use: set ranges = { 4 }.
If you have a fast computer
(or a lot of patience) and want to run multiple
smoothing sizes in the same program, add more to the vector, but it is
unwise.
The alpha_2.5 parameter is set in the beginning of the code. In fact,
this is NOT THE SAME ALPHA_2.5 AS THE PN PAPERS TALK ABOUT!!!
This parameter is what we use to correlate light
with PN, and is set at a "high" value of 100 000 x 10^-9 (whereas if we
were talking about the "real"
alpha_2.5, a normal value would be 23 x 10^-9). This
value is large so that the catalog contains a substantially high number
of PNe so that when we
subsample it down (later) to match the observations,
the subsamples still have appropriate distributions.
Fields are selected as before by the set images = { 7mm.fits }
statement. Again, it is probably unwise to put more than one image in
there. Also, all
images should have the "mm" extension on them, and
there is no distinction at this point between "masked" and "unmasked"
fields - each field only
needs to be run once.
The guts of the program loop over every pixel in the image (that can
have a 9x9 box put around it and still be in the image), do the median
smoothing,
convert that median into a "number of PNe" for that
pixel (using the pretend alpha parameter and flux --> bolometric
luminosity conversion). This
"number" will be greater than 0 and likely not very
large (say, less than 20 or 30). The program takes the decimal part of
that number and treats it as
a probability, drawing a random number and checking
if the decimal part is larger than it, and if it is, then the "number"
gets rounded up, and that is
the number of PNe that go into the catalog at that
pixel.
Example: Say a pixel had a value of 3.1
after the conversions and alpha were applied. Program draws a random
number (say, 0.331) and compares
it with the decimal part
(0.1). As 0.1 !> 0.331, there are 3 PNe here. If the random number
had been less than the decimal part, there would have
been 4 PNe here. This
position is written into the catalog 3 times, and the program moves on
to the next pixel.
Slight modification of late: for fields 7 and Sub,
the code was modified (as mentioned above) to write out the PN
positions for each row in the image.
This allowed it to be distributed over
the cluster and other computers and also prevented memory from getting
upset when you keep working with
vectors that are millions of entries
long. It outputted as many data files as there were rows in the image,
and I concatenated them all into a big file
manually, named with the same
convention as the others.
The positions are written out at the end of the program, into a file
called 8.PNe.3.fits.R4.alpha25100000.RADEC.dat
[RA DEC] where R4 means
smoothing range of 4, or smoothing on 9x9 pixels.
Fields and number of theoretical PNe generated in the master catalog
3
|
4
|
7
|
Core
|
Sub
|
LPC
|
FCJ
|
| 850,978 |
146,918 |
3,863,629 |
1,783,837 |
5,159,820 |
531,963 |
1,007,518 |
The story doesn't end here, though, as a catalog of 1+ million PNe is
not exactly what the analysis code wants to see (or what reality is).
Next up we use
a program called subs.sm to subsample this
massive catalog down to match the observations.
subs.sm reads in the big *RADEC.dat files from each
field and also adds contamination or changes the catalog size, if that
is set:
To set contamination levels, the vector cs
is used - the numbers 0 - 10 are varying contamination from 0% to 100%.
Typically we have been using a
statement like: set cs =
0, 10, 2 to do contamination fractions of 0%, 20%, 40%, 60%, 80%
and 100%.
To change the catalog size, it is done in terms of
the proportion of the observed number. If proportion is set at 100,
then that means that the new catalog
will be100% of the size of the
observed catalog. This has typically been set by set ps = {10 50
100 200 300 500}, for catalogs of 10%, 50%, 100%, 200%, 300%
and 500% of the observed catalogs.
20 separate catalogs are generated by this program, in accordance with
there being traditionally 20 trials of the analysis code run. This is
set in the line:
do itn = 1, 20 {, which runs
everything in the subs.sm
program 20 times, storing the results from each one in a different
catalog file.
These catalog files are written out in pairs, one for the Aguerri
fields and one for the Feldmeier fields (remember the overlap problem)
in files:
PNeFAKE8.f.1.R4.C0.P100.dat
/ PNeFAKE8.a.1.R4.C0.P100.dat
f/a - Feldmeier/Aguerri
1 -
realisation of catalog, 1-20, corresponds with
1-20 trials in the analysis
R4 -
smoothing range on theoretical data generation
(R4 = 9x9 smoothing)
C0 -
contamination fraction (C0 = 0%, C10 = 100%)
P100 -
proportion of observed catalog simulated (P100 =
100%, P10 = 10%, P500 = 500%)
Analysis:
Code for download: PNe26.sm and PNe26Error.sm
In the final result, this analysis is stored in to folder /packmule_raid/steven/PNe26/.
The PNe26.sm file in here
is very similar (intentionally so) to the PNe.sm file
used in the above analysis on the Real Data. The
only difference is the input data file - this version reads in the PNeFAKE8.*.dat file pairs
instead of the single
pair of real data from Feldmeier/Aguerri.
For each trial (1-20, traditionally) of this analysis, a different
subsampled version of the big catalog is used. This is done to allow
this version to get better error bars
and more representative results. If only a single
catalog is subsampled, the "cosmic variance" can be very damaging to
the results. However, I suppose if we wanted
to be really careful, we could generate a different
catalog for each statistics box range within each trial? Or even a new
subsampled catalog everytime we needed one
and never save any of them? (Sounding like our "new"
way to generate theoretical data and do stats... see PNe27 if you're interested.)
PNeError26.sm in this
version is essentially identical to the above version (with the
exception of copying files to PNe26
folder instead of PNe28)
and so it is not discussed.
back to menu
Results of Theoretical Data Analysis:
Well, these results are extremely copious in nature (as there are 6
contamination levels and 6 proportion levels to run (36 combinations)).
To see all the graphs (in their
gorrrrrry detail)
click
here to go to the PNe26 page.
Comparing these two methods:
To see how the various theoretical
data generation results compare to the real data, the two are
overplotted, in different colours. Shown are some example graphs of both
versions plotted together. If you like them, you can
make them again (and any other graph you desire) by going to the re-graphing page.
[still need to make these graphs
once code has run enough]
back to menu
back to main Virgo page
